CSM-lig: small-molecule affinity
CSM-lig: a web server for assessing and comparing protein-small molecule affinities
Douglas E. V. Pires, David B. AscherNucleic Acids Research, v. 44(W1), p. W557-W561, 2016.


Abstract
Interactions between small molecules and proteins mediate many essential biological
processes, and are central in the development of new drugs. Accurately predicting binding affinities is
a challenging and difficult task, and despite being regarded as poorly predictive, scoring functions
play an important role in understanding molecular recognition and in the analysis of molecular
docking results.
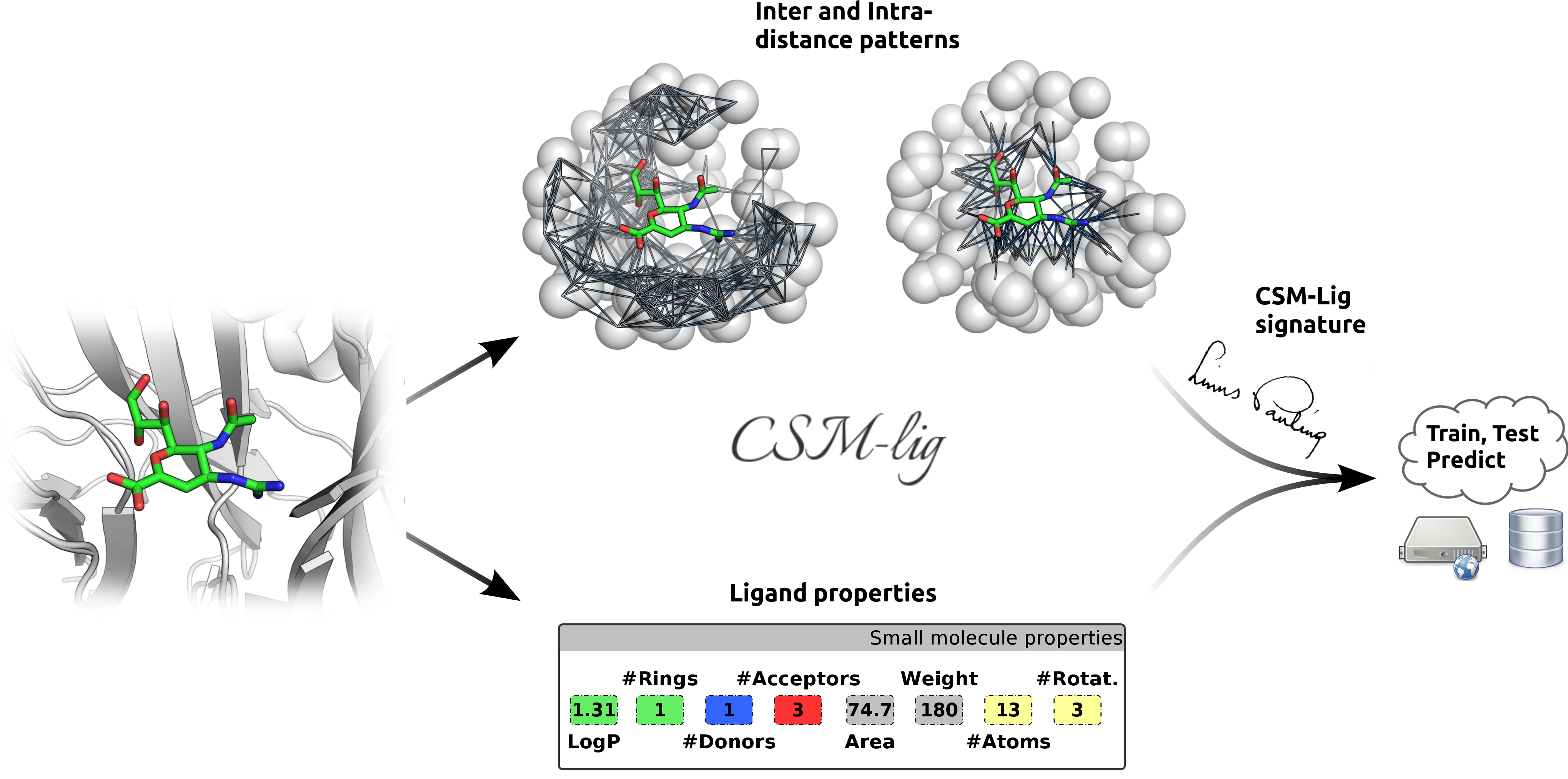
Here we present CSM-Lig,
a web server tailored to predict the binding affinity of a protein-small molecule complex based on
structural signatures. CSM-Lig was first built and evaluated on the widely used 2007 and 2013
releases of PDBbind. For the two releases, CSM-lig achieved a Pearson’s correlation coefficient of
0.82 and 0.86 on 10-fold cross validation. Over the PDBbind core set, a blind test of 195 diverse
complexes with binding affinities ranging from millimolar to picomolar that has been used to
benchmark different approaches, the models showed strong correlations of 0.75 and 0.80,
outperforming well established scoring functions and predictors.
We believe CSM-Lig would be an invaluable tool for helping assess docking poses, the effects
of multiple mutations, including insertions, deletions and alternative splicing events, in protein-small
molecule affinity, unraveling important aspects that drive protein-compound recognition.