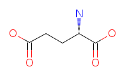
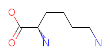| YOUR PROTEIN |
L T C L I G E K D L R L L E K L G D G S F G - V V R R G E - W D A P S G K T V - - - - - - S - V A V K C L K P D V L S - - - - - - - - - - - - - - - - - - - - - - - - - Q P E A M D D F I R E V N A - - M H - - S - - - - L - D - H R N L I R L Y G V V L T P - - - - - - - - - - - - - - P M K M V T E L A P - L G S - L L D - - - - - R L R K H Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - G H F L L G T L S R Y A V Q V A E G M G Y L E S - K - - - - - - - - - - - - R F I H R D L A A R N L L L A T - - - - - - - - - - - - - - - - R D L V K I G D F G L M R A L P Q N D D H Y - - - - - - - - - - - - - - - - - V M Q E H R K V P F A W C A P E S L K T - - - - - - - - - - - - - - - - - - - - - - R T F S H A S D T W M F G V T L W E M F T Y G Q - - - - - - - - - - - - - - - E P W I G L N G S - Q I - - - - - - - - - - - - - - - - - - - - - - - - - - - - L H K I D K E G E R L P - - - - - - - - - R P E D C - - - - - P - - - Q D I - - - Y N V M V Q C W A H K P E D R P T F V A L R D F L L E A Q |
| RAF1_HUMAN |
- - - - - - - - - - - - S T R I G S G S F G - T V Y K G K - W H - - - - - G D - - - - - - - - V A V K I L K V V D P T - - - - - - - - - - - - - - - - - - - - - - - - - - P E Q F Q A F R N E V A V - - L R - - K - - - - T - R - H V N I L L F M G Y M T K D - - - - - - - - - - - - - - N L A I V T Q W C E - G S S - L Y K - - - - - H L H V Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E T K F Q M F Q L I D I A R Q T A Q G M D Y L H A - K - - - - - - - - - - - - N I I H R D M K S N N I F L H E - - - - - - - - - - - - - - - - G L T V K I G D F G L A T V K S R W S G S - - - - - - - - - - - - - - - - - - Q Q V E Q P T G S V L W M A P E V I R M Q D - - - - - - - - - - - - - - - - - - - N N P F S F Q S D V Y S Y G I V L Y E L M T G E - - - - - - - - - - - - - - - - L P Y S H I N N R - D Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - I I F M V G R G Y A S P D L S K L - - - - - Y K N C - - - - - P - - - K A M - - - K R L V A D C V K K V K E E R P L F P Q I L S S I - - - - |
| BRAF_HUMAN |
- - - - - - - - - - - V G Q R I G S G S F G - T V Y K G K - W H - - - - - G D - - - - - - - - V A V K M L N V T A P T - - - - - - - - - - - - - - - - - - - - - - - - - - P Q Q L Q A F K N E V G V - - L R - - K - - - - T - R - H V N I L L F M G Y S T K P - - - - - - - - - - - - - - Q L A I V T Q W C E - G S S - L Y H - - - - - H L H I I - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E T K F E M I K L I D I A R Q T A Q G M D Y L H A - K - - - - - - - - - - - - S I I H R D L K S N N I F L H E - - - - - - - - - - - - - - - - D L T V K I G D F G L A T V K S R W S G S - - - - - - - - - - - - - - - - - - H Q F E Q L S G S I L W M A P E V I R M Q D - - - - - - - - - - - - - - - - - - - K N P Y S F Q S D V Y A F G I V L Y E L M T G Q - - - - - - - - - - - - - - - - L P Y S N I N N R - D Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - I I F M V G R G Y L S P D L S K V - - - - - R S N C - - - - - P - - - K A M - - - K R L M A E C L K K K R D E R P L F P Q I L A S - - - - - |
| GbpC_Ddis |
- - - - - - - - - - - I N K E V G R G A F G - I V Y E A E - W M - - - - - S E - - - - W - - - I A L K K L L L P S S T D T T N L N G N G E V M V - - Y D D P D Q L I E D R L R V F R E F R H E V Y Y - - M S - - G - - - L - - N - H P N V M K I S G F C I Q P - - - - - - - - - - - - - - - L C M A L E Y V R - Y G S - L Y S - - - - - L L S N S - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - S I E I S W G L R L Q I A S E I A K G M Q H L H S - H - - - - - - - - - N P - P V I H R D L K S P N I L L N G I T E G Q - - - - - - - - - - - N S V A T I I D F G T S T A L Y G G - - - - - - - - - - - - - - - - - - - - - A A L I R C V D Q P L W L G P E V L A G - - - - - - - - - - - - - - - - - - - - - - T A Y S E P S D V Y S F G I I L W E L Y T R A - - - - - - - - - - - - - - - - H P F D E F Q F G - Q W M - - - - - - - - - - - - - - - - - - - - - - - - S K - L E D E I I R G L R P T - - - - - - - - - I P P T C - - - - - P - - - P E Y - - - V E L I Q S C W T H E P N S R P T F T S I V E I L G - - - |
| Roco11_Ddis |
- - - - - - - - - - - - E E Q I G V G G F G - L V H K G K - L I - - - - - L Q D K S L V - - - V A I K S Y I V G N S S - - - - - - - - - - - - - - - - - - - - - - A S D I I R K F Q E F H H E M Y I - - M S - - S - - - L - - N - H L N I V K L F G S M Q N P - - - - - - - - - - - - - - - P R M V M E F A P - H G D - L Y H - - - - - F L E K - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K - - K N I K W S F K V R L M L D I A K G I E Y L Q N - Q - - - - - - - - - N P - P I V H R D L R S P N I F L F S L D E N A - - - - - - - - - - - P V C A K V A D F G L S Q Q S L Y S V - - - - - - - - - - - - - - - - - - - - - - - S G L L G N F Q W M A P E T I G A E - - - - - - - - - - - - - - - - - - - - - E S Y T E K I D T Y S F S M I L F T I L T G E - - - - - - - - - - - - - - - - C P F D E F T S F - G K - - - - - - - - - - - - - - - - - - - - - - - - - M E F I R K I R E E D L R P T - - - - - - - - - I P S D C - - - - - P - - - P T I - - - S N L I E L C W S G D P K K R P H F S Y I - - - - - - - - |
| DDB0218878e_Ddis |
- - - - - - - - - - - - G D V I A A G A S G - K V Y K G I - Y K - - - - - G R - - - - D - - - V A I K V Y S S E N F - - - - - - - - - - - - - - - - - - - - - - - - - - - C F N I D E F D R E V T I - - M S - - L - - - I D S D - H P N F T R F Y G A N K Q N K - - - - - - - - - - - - K Y L F H V S E L V K - S G S - L R D - - - - - L L L D K E - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K P L L Y F T Q L S I A S D I A N A M K H L H S - I - - - - - - - - - - - - G V I H R D L K S L N V L I T E D F - - - - - - - - - - - - - - - - T A K V I D F G T S R N V D L A - - - - - - - - - - - - - - - - - - - - - K H M T M N L G T S C Y M S P E L F K G - - - - - - - - - - - - - - - - - - - - - - N G Y D E T C D V Y A F G I V L W E I I A R K - - - - - - - - - - - - - - - - E P Y E N I N S W - S I P - - - - - - - - - - - - - - - - - - - - - - - - - - - - - V M V A K G D R P T - - - - - - - - - I P A D C - - - - - P - - - S E Y - - - S K L I K A C W T D K P K K R P S F K E I C D T L K - - - |
| SgK288_Mmus |
- - - - - - - - - - - - - - - - A S G G F S - K V F Q A R H K R - - - - - W R - - - - - - T Q Y A I K C S P C L Q K E T - - - - - - - - - - - - - - - - - - - - T S S E V T C - - - - L F E E A V K - - M E - - K - - - - I - K - F Q H I V S I Y G V C K - - - - - - - - - - - - - Q P - - L G I V M E F M A - S G S - L E K - - - - - T L P T - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - H S L C W P L K L R I I H E T S L A M N F L H S - I - - - - - - - - - K P - P L L H L D L K P G N I L L D - - - - - - - - - - - - - - - N - N M H V K I S D F G L S K W M E Q S T Q K Q - - - - - - - - - - - - - - - - Y I E R S A L R G T L S Y I P P E M F L E N - - - N - - - - - - - - - - - - - - - - - K A P G P E Y D V Y S F A I V I W E I L T Q K - - - - - - - - - - - - - - - - K P Y A G L N M - - M T - - - - - - - - - - - - - - - - - - - - - - - - - - - - I I I R V A A G M R P S - L Q D V S - D E W - P E E - - - - - V - - - H Q M - - - V N L M K R C W D Q D P K K R P C F L - - - - - - - - - - |
| ARAF_Hsap |
- - - - - - - - - - - L L K R I G T G S F G - T V F R G R - W H - - - - - G D - - - - - - - - V A V K V L K V S Q P T - - - - - - - - - - - - - - - - - - - - - - - - - - A E Q A Q A F K N E M Q V - - L R - - K - - - - T - R - H V N I L L F M G F M T R P - - - - - - - - - - - - - - G F A I I T Q W C E - G S S - L Y H - - - - - H L H V A - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - D T R F D M V Q L I D V A R Q T A Q G M D Y L H A - K - - - - - - - - - - - - N I I H R D L K S N N I F L H E - - - - - - - - - - - - - - - - G L T V K I G D F G L A T V K T R W S G A - - - - - - - - - - - - - - - - - - Q P L E Q P S G S V L W M A A E V I R M Q D - - - - - - - - - - - - - - - - - - - P N P Y S F Q S D V Y A Y G V V L Y E L M T G S - - - - - - - - - - - - - - - - L P Y S H I G C R - D Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - I I F M V G R G Y L S P D L S K I - - - - - S S N C - - - - - P - - - K A M - - - R R L L S D C L K F Q R E E R P L F P Q I L A T - - - - - |
| Roco9_Ddis |
- - - - - - - - - - - - - - - I G E G G T A - T V Y K G K - W R - - - - - Q S - - - - D - - - V A I K L L K T D V V G - - - - - - - - - - - - - - - - - - - - - - - S D F S K V F A E Y R R E I F C - - L S - - S - - - F - - I - H D N I L D L K G F C L E P - - - - - - - - - - - - - - - L A I I T E F Q S - G G N - L Y D - - - - - Y I H D L - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K N P L D W Q L R I K I A K G I A A S L Q T L H D - S - - - - - - - - - R P - S V V H R D L K S P N I L L S S K D - S L - - - - - - - - - - - T M E C H L C D F S L S G F S T T - - - - - - - - - - - - - - - - - - - - - - - V A N R S V Q N P V W L A P E V I N N - - - - - - - - - - - - - - - - - - - - - - E L C S D K S D V Y A Y G V I L F E L L S R T - - - - - - - - - - - - - - - - R F F S N I T F M - S E - - - - - - - - - - - - - - - - - - - - - - - - - - - - V E G L I T E G V R P S - - - - - - - - - L P S H N - - - - - L - - - P E Y - - - D S L L N I C W A Q D P T C R P S F I E I T K K L E - - - |
| BMPR1B_Hsap |
- - - - - - - - - - Q M V K Q I G K G R Y G - E V W M G K - W R - - - - - - - - - - G E - K - V A V K V F F T T E - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E A S W F R E T E I Y Q T V - - L - - - - M - R - H E N I L G F I A A D I K G - - - - - - - - - T G S W T Q L Y L I T D Y H E - N G S - L Y D - - - - - Y L K S T - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - T L D A K S M L K L A Y S S V S G L C H L H T E I F S T Q G - - - - K P - A I A H R D L K S K N I L V K K - - - - - - - - - - - - - - - - N G T C C I A D L G L A V K F I S D T - - - - - - - - - - - - - - - - N E V D I P P N T R V G T K R Y M P P E V L D E S L N R N - - - - - - - - - H - - - - - - F Q - S Y I M A D M Y S F G L I L W E V A R R C V S G - - - - - - G I V E E Y Q L P Y H D L V P S - D P S Y - - - - - - - - - - - - - - - - - - - - - - - E D M R E I V C I K K L R P S - F P N R - - W S - - S - D - - - - - - - - - E C L R Q M G K L M T E C W A H N P A S R L T A L R V K K T - - - - - |
| DDB0231199_Ddis |
- - - - - - - - - - - I L Q F L G E G A L A - E V H K G I - W K - - - - - G K - - - - E - - - V A V K I F N E G S F S - - - - - - - - - - - - - - - - - - - - - - F R - - - - - L E D F L K E V A I - - L G - - L - - - I - - S - H P N L L K L K G A C I A P R S - - - - - - - - - H K S T F M I V T E L M H - K G T - L L E - - - - - V I N K N K - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - P L S L E D I I K Y A L S V A Q G L A Y L H S - - - - - - - - - - - - - V D F I H R D I K A A N I L V D K N - - - - - - - - - - - - - - - - N N A K V G D F G L S R V I D N - - N - - - - - - - - - - - - - - - - - - - F N M T A V A G T P K W E S P E C L M G - - - - - - - - - - - - - - - - - - - - - - E A Y T S A S D V Y S Y G M M L F E L A T G D - - - - - - - - - - - - - - - - E P F L E I Q S I - V E L - - - - - - - - - - - - - - - - - - - - - - - - - A - - R S V C D K K L K P K - - - - - - - - - I S S S V - - - - - P - - - N F I - - - S S L I K D C L H N S P K K R P T M N Q I I Q K L - - - - |
| ILK_Mmus |
- - - - - - - - - - - F L A K L N E N H S G - E L W K G R - W Q - - - - - G N - - - - - - D - I V V K V L K V R D W S T - - - - - - - - - - - - - - - - - - - - - - - - - - R K S R D F N E E C P R - - L R - - I - - - - F - S - H P N V L P V L G A C Q A P - - - - - - - - - - - P A P H P T L I T H W M P - Y G S - L Y N - - - - - V L H E G T - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - N F V V D Q S Q A V K F A L D M A R G M A F L H T - L - - - - - - - - - E P - L I P R H A L N S R S V M I D E D M - - - - - - - - - - - - - - - - T A R I - S M A D V K F S F Q - - - - - - - - - - - - - - - - - - - - - - - - C P G R M Y A P A W V A P E A L Q K K P - - - - - - - - - - - - - - - - - - - E D T N R R S A D M W S F A V L L W E L V T R E - - - - - - - - - - - - - - - - V P F A D L S N M - E I - - - - - - - - - - - - - - - - - - - - - - - - - - - - G M K V A L E G L R P T - I P P G - - - - - - I - - - - - - - S - - - P H V - - - C K L M K I C M N E D P A K R P K F D M I V P I L - - - - |
| HH498_Hsap |
- - - - - - - - - - E F H E I I G S G S F G - K V Y K G R - C R - - - - - N K - - - - I - - - V A I K R Y R A N T Y C - - - - - - - - - - - - - - - - - - - - - - S K - - - S D V D M F C R E V S I - - L C - - Q - - - L - - N - H P C V I Q F V G A C L N D P - - - - - - - - - - - - S Q F A I V T Q Y I S - G G S - L F S - - - - - L L H E Q K - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - R I L D L Q S K L I I A V D V A K G M E Y L H N - L - - - - - - - - - - T Q P I I H R D L N S H N I L L Y E D - - - - - - - - - - - - - - - - G H A V V A D F G E S R F L Q S L D E - - - - - - - - - - - - - - - - - - - D N M T K Q P G N L R W M A P E V F T Q C - - - - - - - - - - - - - - - - - - - - - T R Y T I K A D V F S Y A L C L W E I L T G E - - - - - - - - - - - - - - - - I P F A H L K P A - A A A - - - - - - - - - - - - - - - - - - - - - - - - - A - - - D M A Y H H I R P P - - - - - - - - - I G Y S I - - - - - P - - - K P I - - - S S L L I R G W N A C P E G R P E F S E V V M K L E - - - |
| TESK2_Mmus |
- - - - - - - - - - - - - - K I G S G F F S - E V F K V R - H R - - - - - A S - - - G Q - - - V M A L K M N T - - - - - - - - - - - - - - - - - - - - - - - - - - - - - L S S N R A N L L K E M Q L - - M N - - R - - - L - - S - H P N I L R F M G V C V H Q G - - - - - - - - - - - - - Q L H A L T E Y I N - S G N - L E Q - - - - - L L D S D - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - L Y L P W T V R V K L A Y D I A V G L S Y L H F - K - - - - - - - - - G - - - I F H R D L T S K N C L I K R D - E - N - - - - - - - - - - - G Y S A V V A D F G L A E K I P D A S - - - - - - - - - - - - - - - - I G - - R E K L A V V G S P F W M A P E V L R D - - - - - - - - - - - - - - - - - - - - - - E P Y N E K A D V F S Y G I I L C E I I A R I - - - - - - - - - - - - - - - - Q A D P D Y L P R - T E N - - - - - - - - - - - - - - - - - - - - - - - - F G - - - L D Y D A F Q N - M - - - - - - - - - V - G D C - - - - - P - - - S D F - - - L Q L T F N C C N M D P K L R P S F E E I G K T L - - - - |
| KSR2_Spur |
- - - - - - - - - - - Y G R C I E K G K L G - S T Y K G H - W H - - - - - G E - - - - - - V - L - I Q T R Y - - S K S T - - - - - - - - - - - - - - - - - - - - - - - - - - - D I N D F L D E V A M - - L S - - K - - - - I - R - H E N I A L F M G A C V E P - - - - - - - - - - - G - - N L A V V N S I H K - G P S - L Y H - - - - - H L H V Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - R R K L Q T S S K M D I A R Q I G Q A M G Y L Q A - K - - - - - - - - - - - - G I I V G H L S S R C V Y L E S K V - - - - - - - - - - - - - - - - K L S V T R V D T S D V T C - - - - - Q - - - - - - - - - - - - - - - R A S H A C L P R G K L Q Y L A P E L L A K A R V V P - - - - - - - - P E F - - I T - G N E Q T H S T D V Y A F G T I L F E I I A G R - - - - - - - - - - - - - - - - W P L Q Y A C - S - E S - - - - - - - - - - - - - - - - - - - - - - - - - - - - V I Y Q T C S G R R D D - F R N V - - - - - - T - C - - - - - S - - - S T M - - - K N L I Q E C W S Q N P N E R P S F Q D I V K E L K - - - |
| ank1_Spur |
- - - - - - - - - - - - - - - - G G G S F G - E V F K A V H A V - - - - - H G - - - - - - D - V A V K L A K P G K E R Q - - - - - - - - - - - - - - - - - - - - L E M Y K K Q V V M Y Q K E A Q K H - - E N - - S - - - - L - K - C E H I V P I K G F V Q C E D K - - - - - - A C D C E - L F G I V I Q Y M K - N G S - L W N - - - - - F R T T K W R Q - - - - - - H P D H - - - - - - - - - - - - L D F R T T K L L H H P D L W P L T N R M V Y Q I S R G M H F L H S - K - - - - - - - - - - - - Y I I H R D L K L E N V L V D - - - - - - - - - - - - - - - R - N L N V K I A D L G L A T N L Q T S S - - - - - - - - - - - - - - - - - - - - - - V - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - |
| TNNI3K_Spur |
- - - - - - - - - - E F L E T I G S G S F G - N V Y K G Y - C R - - - - - G K - - - - I - - - V A I K R Y R S S A F S - - - - - - - - - - - - - - - - - - - - - - A K - - - S D V D M F C R E V S I - - L C - - R - - - L - - D - S P Y V I R F V G A C I E D P - - - - - - - - - - - - S H F A I V T Q Y V A - G G S - L F S - - - - - L L H V Q K - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - R N I D L Q S K M T I A V D V A H G M D Y L H N - L - - - - - - - - - - P H P I I H R D L N S H N I L L D E F - - - - - - - - - - - - - - - - G H A E V A D F G E S R F V K S M H E - - - - - - - - - - - - - - - - - - - D N M T K Q P G N L R W M A P E V F S Q N - - - - - - - - - - - - - - - - - - - - - T Q Y S I K A D I F S Y A L C I W E L L S G E - - - - - - - - - - - - - - - - L P F A H L K P A - A A A - - - - - - - - - - - - - - - - - - - - - - - - - A - - - E M A Y R S T R P P - - - - - - - - - I A I T I - - - - - P - - - K S I - - - V N I L Q M M W S P N P E E R P T F A Q I - - - - - - - - |
| Ilk_Dmel |
- - - - - - - - - - - - - - - - - - - - - - - E T W R G R - W Q - - - - - K N - - - - - - D - V V A K I L A V R Q C T P - - - - - - - - - - - - - - - - - - - - - - - - - - R I S R D F N E E F P K - - L R - - I - - - - F - S - H P N I L P I I G A C N S P - - - - - - - - - - - P - - N L V T I S Q F M P - R S S - L F S - - - - - L L H G A T - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - G V V V D T S Q A V S F A L D V A R G M A F L H S - L - - - - - - - - - E R - I I P T Y H L N S H H V M I D D D L - - - - - - - - - - - - - - - - T A R I - N M G D A K F S F Q - - - - - - - - - - - - - - - - - - - - - - - - E K G R I Y Q P A W M S P E T L Q R K Q - - - - - - - - - - - - - - - - - - - A D R N W E A C D M W S F A I L I W E L T T R E - - - - - - - - - - - - - - - - V P F A E W S P M - E C - - - - - - - - - - - - - - - - - - - - - - - - - - - - G M K I A L E G L R V K - I P P G - - - - - - T - - - - - - - S - - - T H M - - - A K L I S I C M N E D P G K R P K F - - - - - - - - - - - |
| TKL6_Spur |
- - - - - - - - - - - - G E T V G R G A F G - E V K K A V - W R - - - - - G T - - - - - - T - V A V K I I S - - T A G S - - - - - - - - - - - - - - - - - - - - - - - - - - - K K D E V E K E V S I - - H K - - R - - - - A - R - H P N I V S L M A V G H R I - - - - - - - - - - - G - - Q V L I V M E F I D - G M N - L H D - - - - - V I F R K K D - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - G - - I T L T P E L K M S I T V G L L S A L T F L H A - S - - - - - - - - - - - - K I L H L D I K P S N V M V E R A S - - - - - - - - - - - - - - - R R A Y L C D L G L A H I K T R S S M - S - - - - - - - - - - - - - - - L S T S L R G P R G T I T H F A P E M V Q K Q G - - - - - - - - - - - - - - - - - - K A - W A G P G N D I W S I A C T F L E L F V E Q - - - - - - - - - - - - - - - - T L W E G V E F L - K L - - - - - - - - - - - - - - - - - - - - - - - - - - - - L R - - T L L K P D A V - P P I L - - - - - - K K V - - - - - K - - - P E V - - - R T I L K P C F D K E P L K R P S A E V L L G - - - - - - |
| HH498_Ddis |
- - - - - - - - - - - T A R K I G A G A F G - D V Y L - V E W R - - - - - N K - - - - - - N - V A V K R V K I E K I L E - - - - - - - - - - - - - - - - - - - - S G K S Y Q W I R D K F I L E A V L - - M V - - K - - - L S - N - F S S F V K L Y A T C I - - - - - - - - - - - - - E E K E L L L V L E F C D - N G S - L Y T - - - - - I L N T I P - - - - - - - - - - - - - - - - - I G G A G A N N N N N N N N N N D I I Q S L P S I N T L S L N I A N G M N Y L H S - L - - - - - - - - - K P - Q I I H R D L T S Q N I L I D - - - - - - - - - - - - - - - R - N G I A K I A D F G I S R F K N D I G D K - - - - - - - - - - - - - - - - - - - - T M T S I G N P R F R S P E V T K G - - - - - - - - - - - - - - - - - - - - - - Q K Y S E K V D V F G F G M I L Y E M F T R R - - - - - - - - - - - - - - - - V P F H D Y E Q I - - A - - - - - - - - - - - - - - - - - - - - - - - - - - - - A S F K I A N A E R P P - L - P Q T - I - - - D - - - - - - - - - - - H R W - - - S N L I Q I C W D Q N P N N R P S F D Q I L T - - - - - - |
| SAPKalpha_Ddis |
- - - - - - - - - - E Y T E K L G A G S S G - K V Y K G L - Y R - - - - - G K - - - - E - - - V A I K V L K S M T - - - - - - - - - - - - - - - - - - - - - - - - - - - E S K E I E E F K K E F Q I - - M S - - A - - - I - - R - S K H V V H F Y G A V L E P - - - - - - - - - - - - - - K L C M V M E N C S - R G S - L Y H - - - - - V M D N N - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - S L D I G W E R T F R F A I E T V R G I E C L H K - W - - - - - - - - - D P - P I V H R D L K S L N L L V N D - - - - - - - - - - - - - - - - K W E I K V C D F G L S R F N T G S N L E - - - - - - - - - - - - - - - - - - - T L V K M R G T F A Y C A P E V Y Y G - - - - - - - - - - - - - - - - - - - - - - E Q F S G K S D V Y S I A V I L W E L V T R C I N G - - - - - - - - - - R Y E R P F S E Y K N L - Q H D - - - - - - - - - - - - - - - - - - - - - - - - F Q I I I Q T A K K N L R P T - - - - - - - - - I P N A C - - - - - P - - - E S L - - - V S L I Q D C W D P N L E N R P T C T D I L S - - - - - - |
| LRRK2_Hsap |
- - - - - - - - - - - - - - - - - D G S F G - S V Y R A A - Y E - - - - - G E - - - - - - E - V A V K I F N K H T - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - S L R L L R Q E L V V - - L C - - H - - - - L - H - H P S L I S L L A A G I R - - - - - - - - - - - - - P - - R M L V M E L A S - K G S - L D R - - - - - L L Q Q D K A - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - S L T R T L Q H R I A L H V A D G L R Y L H S - A - - - - - - - - - - - - M I I Y R D L K P H N V L L F T L Y P N - - - - - - - - - - A - A I I A K I A D Y G I A Q Y C C R M G I - - - - - - - - - - - - - - - - - - - - - - K T S E G T P G F R A P E V A R - - G - - - - - - - - - - - - - - - - - - - N V I Y N Q Q A D V Y S F G L L L Y D I L T T G G - - - - - - - - - - - - - - - R I V E G L K F P - N - - - E F - - - - - - - - - - - - - - - - - - - - - - - - D E L E I Q G K L P D P - V K E Y - - - - - G C A P - - - - - W - - - P M V - - - E K L I K Q C L K E N P Q E R P T S A Q V F - - - - - - - |
| SAPKalpha_C2d_Ddis |
- - - - - - - - - - E Y T E K L G A G S S G - K V Y K G L - Y R - - - - - G K - - - - E - - - V A I K V L K S M T - - - - - - - - - - - - - - - - - - - - - - - - - - - E S K E I E E F K K E F Q I - - M S - - A - - - I - - R - S K H V V H F Y G A V L E P - - - - - - - - - - - - - - K L C M V M E N C S - R G S - L Y H - - - - - V M D N N - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - S L D I G W E R T F R F A I E T V R G I E C L H K - W - - - - - - - - - D P - P I V H R D L K S L N L L V N D - - - - - - - - - - - - - - - - K W E I K V C D F G L S R F N T G S N L E - - - - - - - - - - - - - - - - - - - T L V K M R G T F A Y C A P E V Y Y G - - - - - - - - - - - - - - - - - - - - - - E Q F S G K S D V Y S I A V I L W E L V T R C I N G - - - - - - - - - - R Y E R P F S E Y K N L - Q H D - - - - - - - - - - - - - - - - - - - - - - - - F Q I I I Q T A K K N L R P T - - - - - - - - - I P N A C - - - - - P - - - E S L - - - V S L I Q D C W D P N L E N R P T C T D I L S - - - - - - |
| TKL4_Spur |
- - - - - - - - - - - - - - - V G K G Q F G - V V H R A S - W R - - - - - G T - - - - - - P - V A V K K I S - - M M G S - - - - - - - - - - - - - - - - - - - - - - - - - - - R K D E I E K E V A I - - H R - - R - - - - A - C - H P N I V Q M M A L G Y Q G - - - - - - - - - - - N - - E A F L V M Q F I E - G P S - L H T - - - - - I I F P D R D - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - D V K I P L D W A K K L Q I S R E V L Q A V T F M H A - C - - - - - - - - - - - - N I L H L D I K P A N I L V E S A T - - - - - - - - - - - - - - - L R P Y L C D L G L A H I K S R N L M - S - - - - - - - - - - - - - - - Q - - S S V N M R G T P C F M P P E A L G A T E - - - - - - - - - - - - - - - - - - P G Y R H S N K H D I W S V A G T L V E L Y S M K - - - - - - - - - - - - - - - - R L W G D S M N P - L A - - - - - - - - - - - - - - - - - - - - - - - - - - - - L M A R I F T G Q M G N - P E S L - - - - - - T A V - - - - - D - - - P K V - - - Q K I L E P C F K Q K P E K R P P A S D L L D Q - - - - - |
| ANKRD3_Hsap |
- - - - - - - - - - - - - - K V G S G G F G - Q V Y K V R H V H - - - - - W K - - - - - - T W L A I K C S P S L H V D D - - - - - - - - - - - - - - - - - - - - - - R E R M E - - - - L L E E A K K - - M E - - M - - - - A - K - F R Y I L P V Y G I C R - - - - - - - - - - - - - E P - - V G L V M E Y M E - T G S - L E K - - - - - L L A S - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E P L P W D L R F R I I H E T A V G M N F L H C - M - - - - - - - - - A P - P L L H L D L K P A N I L L D - - - - - - - - - - - - - - - A - H Y H V K I S D F G L A K C N G L S H S - H - - - - - - - - - - - - - - - - D L S M D G L F G T I A Y L P P E R I R E K - - - S - - - - - - - - - - - - - - - - - R L F D T K H D V Y S F A I V I W G V L T Q K - - - - - - - - - - - - - - - - K P F A D E K N I - L H - - - - - - - - - - - - - - - - - - - - - - - - - - - - I M V K V V K G H R P E - L P P V C - R A R - P R A - - - - - C - - - S H L - - - I R L M Q R C W Q G D P R V R P T F Q E I T S E - - - - - |
| MLK4_Hsap |
- - - - - - - - - - E L K E L I G A G G F G - Q V Y R A T - W Q - - - - - G Q - - - - E - - - V A V K A A R Q D P E Q - - - - - - - - - - - - - - - - - - - - - - D A - - A A A A E S V R R E A R L - - F A - - M - - - L - - R - H P N I I E L R G V C L Q Q - - - - - - - - - - - - - P H L C L V L E F A R - G G A - L N R - - - - - A L A A A N A A - - - - - - - - - - - - - - - - - - - - - P D P R A P G P R R A R R I P P H V L V N W A V Q I A R G M L Y L H E - E - - - - - - - - - A F V P I L H R D L K S S N I L L L E K I E H D - - - - - - - D I C - N K T L K I T D F G L A R E W H R - - - - - - - - - - - - - - - - - - - - - - T T K M S T A G T Y A W M A P E V I K S - - - - - - - - - - - - - - - - - - - - - - S L F S K G S D I W S Y G V L L W E L L T G E - - - - - - - - - - - - - - - - V P Y R G I D G L - A V A - - - - - - - - - - - - - - - - - - - - - - - - - Y - - - G V A V N K L T L P - - - - - - - - - I P S T C - - - - - P - - - E P F - - - A K L M K E C W Q Q D P H I R P S F A L I L E Q L - - - - |
| Tak1_Dmel |
- - - - - - - - - - T L R E K V G H G S Y G - V V C K A V - W R - - - - - D K - - - - L - - - V A V K E F F A S A E - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - Q K D I E K E V K Q - - L S - - R - - - V - - K - H P N I I A L H G I S S Y Q Q - - - - - - - - - - - - - A T Y L I M E F A E - G G S - L H N - - - - - F L H G K - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - V - K P A Y S L A H A M S W A R Q C A E G L A Y L H A - M - - - - - - - - - T P K P L I H R D V K P L N L L L T N K - - - - - - - - - - - - - - - G R N L K I C D F G T V A D K S T - - - - - - - - - - - - - - - - - - - - - - - M M T N N R G S A A W M A P E V F E G - - - - - - - - - - - - - - - - - - - - - - S K Y T E K C D I F S W A I V L W E V L S R K - - - - - - - - - - - - - - - - Q P F K G I D N - - A Y T - - - - - - - - - - - - - - - - - - - - - - - - - - - I Q W K I Y K G E R P P - - - - - - - - - L L T T C - - - - - P - - - K R I - - - E D L M T A C W K T V P E D R P S M Q Y I - - - - - - - - |
| RIPK3_Mmus |
- - - - - - - - - - - K L E F V G K G G F G - V V F R A H - H R - - - - - T W - - - - N H D - V A V K I V N S - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K K I S W E V K A - - M V - - N - - - - L - R - N E N V L L L L G V T E D L Q W - - - - - - - - D F V S G Q A L V T R F M E - N G S - L A G - - - - - L L Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - P E C P R P W P L L C R L L Q E V V L G M C Y L H S - L - - - - - - - - - N P - P L L H R D L K P S N I L L D - - - - - - - - - - - - - - - P - E L H A K L A D F G L S T F Q G G S Q - - - - - - - - - - - - - - S G S G S G S G S R D S G G T L A Y L D P E L L F N V - - - - - - - - - - - - - - - - - - - - N L K A S K A S D V Y S F G I L V W A V L A G R E A E - - - - - - - L V D K T S L I R E - T - - - - V C D R - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - Q S R P P - L T E L P P G S P E T P G - - - - - L - - - E K L - - - K E L M I H C W G S Q S E N R P S F Q D C E - - - - - - - |
| 7TMK1_Ddis |
- - - - - - - - - - - F G Q V I G E G Y F G - E V R K A V - W K - - - - - G A - - - - V - - - V A V K I L H R N S F R - - - - - - - - - - - - - - - - - - - - - - N T D G N K E E N V F L K E V A I - - L S - - I - - - L - - R - H P N V L Q F L G V C S E T - - - - - - - - - - - - - N L N G I V T E Y M G - G G S - L D R - - - - - L L T D R Y - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - F L I K Q N P I L A W N M A I S I A R G M F Y L H D - W - - - - - - - - - K P N P I L H R D L S T K N I L L D E S - - - - - - - - - - - - - - - L T I A K V A D F G L S K E Q G - - - - - - - - - - - - - - - - - - - - - - - F E M T S T V G H L C Y Q A P E V F I G - - - - - - - - - - - - - - - - - - - - - - E L Y T P K A D V Y S F G L L V W C I I T G E - - - - - - - - - - - - - - - - Q P N Q N L Q P L - K M A - - - - - - - - - - - - - - - - - - - - - - - - - H - - - L A A Y E N Y R P P - - - - - - - - - M P Q P M - - - - - D - - - P M W E N L G K L I E M C W K K S P E E R P S F S F I L D F L - - - - |
| wit_Dmel |
- - - - - - - - - - - - I G M L G S G K Y G - T V M K G L - L H - - - - - D Q - - - - - - E - V A V K I Y P E E H - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - H Q Y Y V N E R N I Y A L P - - L - - - - M - E - C P A L L S Y F G Y D E R C T M - - - - - - - - D G R M E Y Q L V L S L A P - L G C - L Q D - - - - - W L I A N - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - T L T F S E C C G M L R S I T R G I S H L H T - E L R L G D - - Q H K P - C V A H R D I N T R N V L V Q - - - - - - - - - - - - - - - A - D L S C C I A D F G F A L K V F G S K Y E Y - K - - - - - - - - G E V A M A E T K S I N E V G T L R Y M A P E L L E G A V N L R - - - - - - - - - - - D - - - - C E T S L K Q M D V Y A L G L V L W E V A T R C S D F - - - - - - - Y A P - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - |
| Gdt9_Ddis |
- - - - - - - - - - - F D E I C G Q G T Y G - M V Y K G K - W R - - - - - S Q - - - - - - V - V A I K M M R - - V G T V - - - - - - - - - - - - - - - - - - - - - - - - - - - N C D E V Y R E M D I - - M G - - K - - - - I - R - H Q N V I S F I G C V V S L - - - - - - - - - - - E - - H L C I V T E F S P - I G S - L G D - - - - - L I H G D N N Q - - - - - - K K S N T N T I T N T I T N T N T N T N T S T S T S T K L T V K Q K L R I A I D I A R G C Q F L H Q - C - - - - - - - - - - - - G I I H R D L K P D N V L V F D I N H N - - - - - - - - - - A - P V C A K I S D F G T S K E T N E F S - - - - - - - - - - - - - - - - - - - - N K N T N C V G T P I Y M S N E I L E K K - - - - - - - - - - - - - - - - - - - - - - T Y D N S T D V Y S F A V L F Y E M M I E E - - - - - - - - - - - - - - - - V P F S E I D E K - W E - - - - - - - - - - - - - - - - - - - - - - - - - - - - I P S K V I S G W R P T - K G L N - - - - - - S - L - - - - - D - - - R D I - - - K N L I N I C W A - - P H S L P S F D E I V - - - - - - - |
| BMPR1A_Mmus |
- - - - - - - - - - - - V R Q V G K G R Y G - E V W M G K - W R - - - - - - - - - - G E - K - V A V K V F F T T E - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E A S W F R E T E I Y Q T V - - L - - - - M - R - H E N I L G F I A A D I K G - - - - - - - - - T G S W T Q L Y L I T D Y H E - N G S - L Y D - - - - - F L K C A - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - T L D T R A L L K L A Y S A A C G L C H L H T E I Y G T Q G - - - - K P - A I A H R D L K S K N I L I K K - - - - - - - - - - - - - - - - N G S C C I A D L G L A V K F N S D T - - - - - - - - - - - - - - - - N E V D I P L N T R V G T K R Y M A P E V L D E S L N K N - - - - - - - - - H - - - - - - F Q - P Y I M A D I Y S F G L I I W E M A R R C I T G - - - - - - G I V E E Y Q L P Y Y N M V P S - D P S Y - - - - - - - - - - - - - - - - - - - - - - - E D M R E V V C V K R L R P I - V S N R - - W N - - S - D - - - - - - - - - E C L R A V L K L M S E C W A H N P A S R L T A L R I K K T - - - - - |
| BMPR2_Hsap |
- - - - - - - - - - - - L E L I G R G R Y G - A V Y K G S - L D - - - - - - - - - - E R - P - V A V K V F S F A N - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - R Q N F I N E K N I Y R V P - - L - - - - M - E - H D N I A R F I V G D E R V - - - - - - - - T A D G R M E Y L L V M E Y Y P - N G S - L C K - - - - - Y L S L H - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - T S D W V S S C R L A H S V T R G L A Y L H T E L P R G D H - - - Y K P - A I S H R D L N S R N V L V K N - - - - - - - - - - - - - - - - D G T C V I S D F G L S M R L T G N R L V R - - - - - - - - - - - - P G E E D N A A I S E V G T I R Y M A P E V L E G A V N L R - - - - - - - - - D - - - - - - C E S A L K Q V D M Y A L G L I Y W E I F M R C T D L F P G - - - E S V P E Y Q M A F Q T E V G N - H P T F - - - - - - - - - - - - - - - - - - - - - - - E D M Q V L V S R E K Q R P K - F P E A - - W K - - E - N - - - - - S - - - L A V R S L K E T I E D C W D Q D A E A R L T A Q C A - - - - - - - - |
| MISR2_Mmus |
- - - - - - - - - - - F S Q V I Q E G G H A - V V W A G R - L Q - - - - - G E - - - - - - M - V A I K A F P P R A - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - V A Q F R A E G A V Y Q L L - - G - - - - L - Q - H D H I V R F I T A G Q G G P G - - - - - - - - P L P S G P L L V L E L Y P - K G S - L C H - - - - - Y L T Q Y - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - T S D W G S S L R M A L S L A E G L A F L H E - E R W Q D G - - Q Y K P - G I A H R D L S S Q N V L I R - - - - - - - - - - - - - - - E - D R S C A I G D L G L A L V L P G L A Q P P A L - - - - - - - - A P T Q P R G P A A I L E A G T Q R Y M A P E L L D K T L D L Q - - - - - - - - - - - D - - - - W G T A L Q R A D V Y S L A L L L W E I L S R C S D L - - - - - - - R P D H R P P P F Q L A Y E A - E L G S N P - - - - - - S - - - - - - - - - - - - - A C E L W A L A V E E R K R P N - I P S T - - W S C S A T D - - - - - P - - - R G L - - - R E L L E D C W D A D P E A R L T A E C V Q Q R L - - - - |
| DDB0231197_Ddis |
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - N - - - - - G M - - - - D - - - V A I K E F S Q D G M - - - - - - - - - - - - - - - - - - - - - - - - - - - G F D W V S F Y K E I T I - - V S - - A - - - S - - Q - H P K V I K C Y G A H T K N T - - - - - - - - - - - - N K P F I V T E L C S - R G T - I S N - - - - - A L N Q I R - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K T T G Q P P A I P L I V H M A I D A A Q S L E F I H S - K - - - - - - - - - - - - N I I H R D V K G N N F L V N E N W - - - - - - - - - - - - - - - - E V K L I D F G V S R F V E A R - - - - - - - - - - - - - - - - - - - - - - L G Y T I V G T P N Y M A C E L F N G - - - - - - - - - - - - - - - - - - - - - - Q P Y H Q P A D V F S F G V V L W E M F T Q D - - - - - - - - - - - - - - - - T P Y K N L T R I - E Q A - - - - - - - - - - - - - - - - - - - - - - - - - - - - - L F V Q S G G R P T - - - - - - - - - I P P T V - - - - - P - - - T T I - - - A N L I E S C W V Q T P H L R P T F T E I L K V - - - - - |
| BRAF_Hsap |
- - - - - - - - - - - V G Q R I G S G S F G - T V Y K G K - W H - - - - - G D - - - - - - - - V A V K M L N V T A P T - - - - - - - - - - - - - - - - - - - - - - - - - - P Q Q L Q A F K N E V G V - - L R - - K - - - - T - R - H V N I L L F M G Y S T K P - - - - - - - - - - - - - - Q L A I V T Q W C E - G S S - L Y H - - - - - H L H I I - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E T K F E M I K L I D I A R Q T A Q G M D Y L H A - K - - - - - - - - - - - - S I I H R D L K S N N I F L H E - - - - - - - - - - - - - - - - D L T V K I G D F G L A T V K S R W S G S - - - - - - - - - - - - - - - - - - H Q F E Q L S G S I L W M A P E V I R M Q D - - - - - - - - - - - - - - - - - - - K N P Y S F Q S D V Y A F G I V L Y E L M T G Q - - - - - - - - - - - - - - - - L P Y S N I N N R - D Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - I I F M V G R G Y L S P D L S K V - - - - - R S N C - - - - - P - - - K A M - - - K R L M A E C L K K K R D E R P L F P Q I L A S - - - - - |
| MLK3_Mmus |
- - - - - - - - - - R L E E V I G I G G F G - K V Y R G S - W R - - - - - G E - - - - L - - - V A V K A A R Q D P D E - - - - - - - - - - - - - - - - - - - - - - D I - - S V T A E S V R Q E A R L - - F A - - M - - - L - - A - H P N I I A L K A V C L E E - - - - - - - - - - - - - P N L C L V M E Y A A - G G P - L S R - - - - - A L A G - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - R R V P P H V L V N W A V Q I A R G M H Y L H C - E - - - - - - - - - A L V P V I H R D L K S N N I L L L Q P I E G D - - - - - - - D M E - H K T L K I T D F G L A R E W H K - - - - - - - - - - - - - - - - - - - - - - T T Q M S A A G T Y A W M A P E V I K A - - - - - - - - - - - - - - - - - - - - - - S T F S K G S D V W S F G V L L W E L L T G E - - - - - - - - - - - - - - - - V P Y R G I D C L - A V A - - - - - - - - - - - - - - - - - - - - - - - - - Y - - - G V A V N K L T L P - - - - - - - - - I P S T C - - - - - P - - - E P F - - - A Q L M A D C W A Q D P H R R P D F A S I L Q Q L - - - - |
| ZAK_Hsap |
- - - - - - - - - - - F F E N C G G G S F G - S V Y R A K - W I - - - - - S Q - - - - D K E - V A V K K L L - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K I E K E A E I - - L S - - V - - - L - - S - H R N I I Q F Y G V I L E P - - - - - - - - - - - - - P N Y G I V T E Y A S - L G S - L Y D - - - - - Y I N S - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - N R S E E M D M D H I M T W A T D V A K G M H Y L H M - E - - - - - - - - - A P V K V I H R D L K S R N V V I A A - - - - - - - - - - - - - - - - D G V L K I C D F G A S R F H N H - - - - - - - - - - - - - - - - - - - - - - T T H M S L V G T F P W M A P E V I Q S - - - - - - - - - - - - - - - - - - - - - - L P V S E T C D T Y S Y G V V L W E M L T R E - - - - - - - - - - - - - - - - V P F K G L E G L - Q V A - - - - - - - - - - - - - - - - - - - - - - - - - W - - - L V V E K N E R L T - - - - - - - - - I P S S C - - - - - P - - - R S F - - - A E L L H Q C W E A D A K K R P S F K Q I I S I - - - - - |
| DDB0229872_Ddis |
- - - - - - - - - - T V E K E I G Q G F F G - K V Y K A R - W R - - - - - G K - - - - S - - - V A L K K I T L I K F R - - - - - - - - - - - - - - - - - - - - - - D L - - - T E T E I F D K E V S I - - M S - - K - - - L - - C - H P T C V M F I G A C S L D G - - - - - - - - - - P S N D R S I I M E Y M E - G G S - L R R - - - - - L L D E K S - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - S Y H L P P S L Q L S I A R D I A E G M N Y L H T N F - - - - - - - - - K E G P I V H R D L T S S N I L L N S S - - - - - - - - - - - - - - - Y T V A K I N D F G L S K E M K P - G P - - - - - - - - - - - - - - - - - - - T E M T A A M G S L A W M A P E C F K A - - - - - - - - - - - - - - - - - - - - - - E N Y T E K V D V Y S F A I I L W E I V T C R - - - - - - - - - - - - - - - - D P Y N G M E P L - R L A - - - - - - - - - - - - - - - - - - - - - - - - - F - - - L A S V E D Y R L P - - - - - - - - - L N G - F - - - - - P - - - P Y W V - - - E L I S K C W N I T P S L R P S F K E I L Q I - - - - - |
| ALK1_Hsap |
- - - - - - - - - - - - V E C V G K G R Y G - E V W R G L - W H - - - - - - - - - - G E - S - V A V K I F S S R D - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E Q S W F R E T E I Y N T V - - L - - - - L - R - H D N I L G F I A S D M T S - - - - - - - - - R N S S T Q L W L I T H Y H E - H G S - L Y D - - - - - F L Q R Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - T L E P H L A L R L A V S A A C G L A H L H V E I F G T Q G - - - - K P - A I A H R D F K S R N V L V K S - - - - - - - - - - - - - - - - N L Q C C I A D L G L A V M H S Q G S - - - - - - - - - - - - - - - - D Y L D I G N N P R V G T K R Y M A P E V L D E Q I R T D - - - - - - - - - C - - - - - - F E - S Y K W T D I W A F G L V L W E I A R R T I V N - - - - - - G I V E D Y R P P F Y D V V P N - D P S F - - - - - - - - - - - - - - - - - - - - - - - E D M K K V V C V D Q Q T P T - I P N R - - L A - - A - D - - - - - - - - - P V L S G L A Q M M R E C W Y P N P S A R L T A L R I K K T L - - - - |
| MLK2_Mmus |
- - - - - - - - - - Q L E E I I G V G G F G - K V Y R A V - W R - - - - - G E - - - - E - - - V A V K A A R L D P E R - - - - - - - - - - - - - - - - - - - - - - D P - - A V T A E Q V R Q E A R L - - F G - - A - - - L - - Q - H P N I I A L R G A C L S P - - - - - - - - - - - - - P N L C L V M E Y A R - G G A - L S R - - - - - V L A G - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - R R V P P H V L V N W A V Q V A R G M N Y L H N - D - - - - - - - - - A P V P I I H R D L K S I N I L I L E A I E N H - - - - - - - N L A - D T V L K I T D F G L A R E W H K - - - - - - - - - - - - - - - - - - - - - - T T K M S A A G T Y A W M A P E V I R L - - - - - - - - - - - - - - - - - - - - - - S L F S K S S D V W S F G V L L W E L L T G E - - - - - - - - - - - - - - - - V P Y R E I D A L - A V A - - - - - - - - - - - - - - - - - - - - - - - - - Y - - - G V A M N K L T L P - - - - - - - - - I P S T C - - - - - P - - - E P F - - - A R L L E E C W D P D P H G R P D F G S I L K Q L E - - - |
| Tesk_Spur |
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - V M V A K I G K Q K I - - - - - - - - - - - - - - - - - - - - - - - - - - W R A N Q H K M I Q E L E L - - L N - - K - - - L - - Q - H P N V V R Y M G A C V K D G - - - - - - - - - - - - - H I H P V L E Y V S - G G C - L T D - - - - - I L A D E - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - S L A L S W R Q K G D L A T D I A R G M T Y L H S - Q - - - - - - - - - N - - - V C H R D L T S A N C L V R Q K - P N N - - - - - - - - - - - V L E A I L T D F G L A R V L G - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - |
| Roco10_Ddis |
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - S F R H E A L S - - L Y - - N - - - - F - R - H S N V L S I I G I S M - - - - - - - - - - - - - N - - P I A I I T E N P T - F G S - K G S T T L S E Y I Q D R - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - Q K H P E I P W N I K L K I A L D I A R A M D K L N S - H - - - - - - - - - S P - P F L L T N L T S S G I I L E K S N D H Q S S G N G L E I E P - F V N A K I I D F S Q S S F L P S L - - - - - - - - - - - - - - - - - - - - - - - F P T S T T P K S Y H S P E V L Q K L - - - - - - - - - - - - - - - - - - - - - - N Y H E N S D V Y S F G I I L Y E L L T R S - - - - - - - - - - - - - - - - I A F Q D H D - - - - R - - - - - - - - - - - - - - - - - - - - - - - - - - - - F S N I V I S G Q R P S - I P L D - - - - - - C - - - - - - - L - - - P S F - - - A D L I K D C W S G E P L N R P S P S K I I S - - - - - - |
| DDB0230124_Ddis |
- - - - - - - - - - - V G S R L G L G S F G - D C Y T - G N A F - - - - - S I - - - - - - P - T I L K K L R T Q R F T D - - - - - - - - - - - - - - - - - - - - Q - - - - - - F L G Q F K N E V M Q - - I R - - E - - - - L - Q - H E N L V P V S G C C L - - - - - - - - - - - - - D N - N I Y I V H P Y Y D - A S N - L Q T - - - - - L L F E Q S H N - - - - - - N N N N N - N N N N N N N N N N N N N N N N N N N H T I I S N E F I H R V S L G I S K A M T Y L H S - H - - - - - - - - - - - - D V V H R A L K L N N I L I D - - - - - - - - - - - - - - - R V S G N V L V R D Y G F N F V K D G I F - - - - - - - - - - - - - - - - - - - - - - - R T G Q Q S S P Y L A P E L F N S - - - N C - - - - - - - - - - - - - - - - - N Q Y D T N S D Q F G F A M I L L Q L F T R S - - - - - - - - - - - - - - - - P L F Q D I H V S - - R - - - - - - - - - - - - - - - - - - - - - - - - - - - - I T D T I L N G V R P E - I - P D N - V - - - P - - - - - - - - - - - S V F - - - S R L I K A C W S A D S S A R P S F L T I S K I L - - - - |
| TGFBR2_Mmus |
- - - - - - - - - - - L D T L V G K G R F A - E V Y K A K - L K Q N T S E - - - - Q F E - T - V A V K I F P Y E E - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - Y S S W K T E K D I F S D I - - N - - - - L - K - H E N I L Q F L T A E E R K - - - - - - - - - T E L G K Q Y W L I T A F H A - K G N - L Q E - - - - - Y L T R H - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - V I S W E D L R K L G S S L A R G I A H L H S D H - T P C G - - R P K M - P I V H R D L K S S N I L V K N - - - - - - - - - - - - - - - - D L T C C L C D F G L S L R L D P T L - - - - - - - - - - - - - - - - S V D D L A N S G Q V G T A R Y M A P E V L E S R M N L E - - - - - - - - - N - - - - - - V E - S F K Q T D V Y S M A L V L W E M T S R C N A V - - - - - - G E V K D Y E P P F G S K V R E - H P C V - - - - - - - - - - - - - - - - - - - - - - - E S M K D S V L R D R G R P E - I P S F - - W L - - N - H - - - - - - - - - Q G I Q I V C E T L T E C W D H D P E A R L T A Q C V A E R - - - - - |
| cdi_Dmel |
- - - - - - - - - - - - - E K I G S G F F S - E V Y K V T - H R - - - - - T T - - - G Q - - - V M V L K M N Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - - L R A N R P N M L R E V Q L - - L N - - K - - - L - - S - H A N I L S F M G V C V Q E G - - - - - - - - - - - - - Q L H A L T E Y I N - G G S - L E Q - - - - - L L A N K - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E V V L S A T Q K I R L A L G I A R G M S Y V H D - A - - - - - - - - - G - - - I F H R D L T S K N V L I R N L - A N D - - - - - - - - - - - Q Y E A V V G D F G L A A K I P V K S - - - - - - - - - - - - - - - - R - - - K S R L E T V G S P Y W V S P E C L K G - - - - - - - - - - - - - - - - - - - - - - Q W Y D Q T S D V F S F G I I Q C E I I A R I - - - - - - - - - - - - - - - - E A D P D M M P R - T A S - - - - - - - - - - - - - - - - - - - - - - - - F G - - - L D Y L A F V E - L - - - - - - - - - C P M D T - - - - - P - - - P V F - - - L R L A F Y C C L Y D A K S R P T F H D A T K K L T - - - |
| 26581_Tthe |
- - - - - - - - - - E I Q N K I T E G G Y G - I I Y K A K - W R - - - - - E I - - - - V - - - V A V K K F K I D Y N - - - - - - - - - - - - - - - - - - - - - - - - - - N Q Q Q I V E F V N E C N A - - M E - - A - - - L - - R - H P N I V L F L G A C T E I P - - - - - - - - - - - - - N F S I V M E Y C Q - R G S - L W S - - - - - L L Q N Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - S V P L T W E D R R K I A L D I A K G V F F L H S - S - - - - - - - - - K P - P I I H R D L K S L N V L V D D - - - - - - - - - - - - - - - - N F R C K L T D F G W T R V K P Q D - - - - - - - - - - - - - - - - - - - - - N Y M T N K I G T Y Q W M A P E V I K A - - - - - - - - - - - - - - - - - - - - - - F Y Y T E K A D V F S Y S I I L W E I A S R E - - - - - - - - - - - - - - - - P P Y R G I K G D - V V - - - - - - - - - - - - - - - - - - - - - - - - - - - - A E K V M - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - |
| LRRKc_Spur |
- - - - - - - - - - - - - Q S L G Q G G F G - E V F R A K - F R - - - - - G E - - - - - - T - V A A K T M F P S R L L K - - - - - - - - - - - - - - - - - - - - - - N - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - |
| TESK2_Hsap |
- - - - - - - - - - - - - - K I G S G F F S - E V F K V R - H R - - - - - A S - - - G Q - - - V M A L K M N T - - - - - - - - - - - - - - - - - - - - - - - - - - - - - L S S N R A N M L K E V Q L - - M N - - R - - - L - - S - H P N I L R F M G V C V H Q G - - - - - - - - - - - - - Q L H A L T E Y I N - S G N - L E Q - - - - - L L D S N - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - L H L P W T V R V K L A Y D I A V G L S Y L H F - K - - - - - - - - - G - - - I F H R D L T S K N C L I K R D - E - N - - - - - - - - - - - G Y S A V V A D F G L A E K I P D V S - - - - - - - - - - - - - - - - M G - - S E K L A V V G S P F W M A P E V L R D - - - - - - - - - - - - - - - - - - - - - - E P Y N E K A D V F S Y G I I L C E I I A R I - - - - - - - - - - - - - - - - Q A D P D Y L P R - T E N - - - - - - - - - - - - - - - - - - - - - - - - F G - - - L D Y D A F Q H - M - - - - - - - - - V - G D C - - - - - P - - - P D F - - - L Q L T F N C C N M D P K L R P S F V E I G K T L - - - - |
| KSR1_Hsap |
- - - - - - - - - - - L G E P I G Q G R W G - R V H R G R - W H - - - - - G E - - - - - - - - V A I R L L E M D G H - - - - - - - - - - - - - - - - - - - - - - - - - - N Q D H L K L F K K E V M N - - Y R - - Q - - - - T - R - H E N V V L F M G A C M N P - - - - - - - - - - - - - P H L A I I T S F C K - G R T - L H S - - - - - F V R D P - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K T S L D I N K T R Q I A Q E I I K G M G Y L H A - K - - - - - - - - - - - - G I V H K D L K S K N V F Y D - - - - - - - - - - - - - - - - - N G K V V I T D F G L F G I S G V V R E - - - - - - - - - - - - - - - G R R E N Q L K L S H D W L C Y L A P E I V R E M T P G K - - - - - - - - D - - - - - E D Q L P F S K A A D V Y A F G T V W Y E L Q A R D - - - - - - - - - - - - - - - - W P L K N Q A A E - A S I - - - - - - - - - - - - - - - - - - - - - - - - - - - W Q I G S G E G M K R V - L T S V S - - - - - - - L - - - - - G - - - K E V - - - S E I L S A C W A F D L Q E R P S F S L L M D - - - - - - |
| KinX_Ddis |
- - - - - - - - - - - F I S E I G S G G F G - K V F K G E - Y L - - - - - G A - - - - P - - - V A I K K I H I L P D - - - - - - - - - - - - - - - - - - - - - - - D P N R V D L E K F L N R E I E T - - I K - - L - - - F - - T - H P N V I Q F V G I S E N - - - - - - - - - - - - - N G I L F I V T E L I E - G G D - L Q Y - - - - - Y L K N Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - S I D L P W F L R A N I A L D V S L A M S Y L H S - K - - - - - - - - - - - - S I V H R D L K S T N L L V D K N - - - - - - - - - - - - - - - - W K I K V C D F G F A R I V E E D N - - - - - - - - - - - - - - - - - - - - N K S M T I C G T D N W M S P E M I T G - - - - - - - - - - - - - - - - - - - - - - L D Y D E R S D I F S F G I V L L E I I S R V - - - - - - - - - - - - - - - - K P A P Y M R D A - S - - - - - - - - - - - - - - - - - - - - - - - - - - - - - F G L A E D I V R N Q L - - - - - - - - - I P T D C - - - - - P - - - E S L - - - I D L T F N C C S V D P N N R P S F K E I S Q T L - - - - |
| ACTR2_Mmus |
- - - - - - - - - - - - - - - - - - G R F G - C V W K A Q - L L - - - - - - - - - - N E - Y - V A V K I F P I Q D - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K Q S W Q N E Y E V Y S L P - - G - - - - M - K - H E N I L Q F I G A E K R G - - - - - - - - - T S V D V D L W L I T A F H E - K G S - L S D - - - - - F L K A N - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - V V S W N E L C H I A E T M A R G L A Y L H E D I P G L - K - D G H K P - A I S H R D I K S K N V L L K N - - - - - - - - - - - - - - - - N L T A C I A D F G L A L K F E A G K - - - - - - - - - - - - - - - - S A G - - D T H G Q V G T R R Y M A P E V L E G A I N F Q - - - - - - - - - R - - - - - - - D - A F L R I D M Y A M G L V L W E L A S R C T A A - - - - - D G P V D E Y M L P F E E E I G Q - H P S L - - - - - - - - - - - - - - - - - - - - - - - E D M Q E V V V H K K K R P V - L R D Y - - W Q - - K - H - - - - - - - - - A G M A M L C E T I E E C W D H D A E A R L S A - - - - - - - - - - - |
| DPYK3~b_Ddis |
- - - - - - - - - - - - V E K V G A G S F A - N V F L G I - W N - - - - - G Y - - - - K - - - V A I K I L K N E S - - - - - - - - - - - - - - - - - - - - - - - - - - - - I S N D E K F I K E V S S - - L I - - K - - - S - - H - H P N V V T F M G A C I D P - - - - - - - - - - - - - - - P C I F T E Y L Q - G G S - L Y D - - - - - V L H I Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K I K L N P L M M Y K M I H D L S L G M E H L H S - I - - - - - - - - - Q - - - M L H R D L T S K N I L L D E - - - - - - - - - - - - - - - - F K N I K I A D F G L A T T L S D D - - - - - - - - - - - - - - - - - - - - - - M T L S G I T N P R W R S P E L T K G L - - - - - - - - - - - - - - - - - - - - - - V Y N E K V D V Y S F G L V V Y E I Y T G K - - - - - - - - - - - - - - - - I P F E G L D G T - A S - - - - - - - - - - - - - - - - - - - - - - - - - - - - A A K A A F E N Y R P A - - - - - - - - - I P P D C - - - - - P - - - V S L - - - R K L I T K C W A S D P S Q R P S F T E I L T E L E - - - |
| DDB0229940_Ddis |
- - - - - - - - - - - F G D V I A S G A S G - K V Y K G I - Y K - - - - - G R - - - - D - - - V A I K V Y S S E N F - - - - - - - - - - - - - - - - - - - - - - - - - - - C F N I E E F D R E V T I - - M S - - L - - - I D S D - H P N F T R F Y G A N K Q N K - - - - - - - - - - - - K Y L F H V S E L V K - S G S - L R D - - - - - L L L D K E - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K P L A Y F T Q L S I A S D I A N A M K H L H S - I - - - - - - - - - - - - G V I H R D L K S L N V L I T E D F - - - - - - - - - - - - - - - - T A K V I D F G T S R N V D L A - - - - - - - - - - - - - - - - - - - - - K Q M T L N L G T S C Y M S P E L F K G - - - - - - - - - - - - - - - - - - - - - - N G Y D E T C D V Y A F G I V L W E I I A R K - - - - - - - - - - - - - - - - E P Y E N I N S W - S I P - - - - - - - - - - - - - - - - - - - - - - - - - - - - - V L V A K G E R P T - - - - - - - - - I P A D C - - - - - P - - - S E Y - - - S K L I K A C W T D K P K K R P S F K E I - - - - - - - - |
| ILK_Hsap |
- - - - - - - - - - - F L T K L N E N H S G - E L W K G R - W Q - - - - - G N - - - - - - D - I V V K V L K V R D W S T - - - - - - - - - - - - - - - - - - - - - - - - - - R K S R D F N E E C P R - - L R - - I - - - - F - S - H P N V L P V L G A C Q S P - - - - - - - - - - - P A P H P T L I T H W M P - Y G S - L Y N - - - - - V L H E G T - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - N F V V D Q S Q A V K F A L D M A R G M A F L H T - L - - - - - - - - - E P - L I P R H A L N S R S V M I D E D M - - - - - - - - - - - - - - - - T A R I - S M A D V K F S F Q - - - - - - - - - - - - - - - - - - - - - - - - C P G R M Y A P A W V A P E A L Q K K P - - - - - - - - - - - - - - - - - - - E D T N R R S A D M W S F A V L L W E L V T R E - - - - - - - - - - - - - - - - V P F A D L S N M - E I - - - - - - - - - - - - - - - - - - - - - - - - - - - - G M K V A L E G L R P T - I P P G - - - - - - I - - - - - - - S - - - P H V - - - C K L M K I C M N E D P A K R P K F D M I V P I L - - - - |
| LRRKd_Spur |
- - - - - - - - - - - - - Q S L G Q G G F G - E V F R A K - F R - - - - - G E - - - - T - - - V A A K T M L P S R L L - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - |
| Shk3_Ddis |
- - - - - - - - - - - F E E L I G T G S F G - K V Y K G R - C R - - - - - Q K - - - - A - - - V A V K L L H K Q N F - - - - - - - - - - - - - - - - - - - - - - - - - - D A A T L S A F R K E V H L - - M S - - K - - - I Y - - - H P N I C L F M G A C T I P G - - - - - - - - - - - - - R C V I V T E L V P - K G N - L E T - - - - - L L H D Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K I Q L P L Y L R M R M A R D A A L G I N W L H E - S - - - - - - - - - N P - V F V H R D I K S S N L L V D E - - - - - - - - - - - - - - - - N M R V K I C D F G L S A L K Q K H K M - - - - - - - - - - - - - - - - - - L K D Q S S A K G T P L Y M A P E V M M F - - - - - - - - - - - - - - - - - - - - - - K E F N E S S D V Y S F G I V L W E I L T R K - - - - - - - - - - - - - - - - E P F S H H R E - - L - - - - - - - - - - - - - - - - - - - - - - - - - - E K F R E A V C V K H E R P P - - - - - - - - - I P N D C - - - - - L - - - D S L - - - R R L I E K C W D K E P I S R P S F K E I I S A L D - - - |
| DDB0229865_Ddis |
- - - - - - - - - - - - - - - - - - G S F A - K V Y K G S - Y Y - - - - - G N - - - - P - - - V C V K V I K K D T L - - - - - - - - - - - - - - - - - - - - - - - - - I D K E S W V F L K R E I G I - - L K - - N L L N Q - - G - H K N I I R F I G I G E K - - - - - - - - - - - - - D S L L F L V T E L I N - G G D - L G N - - - - - I L L D H - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K F H I P W S L R V K I A K D I A E G M E Y L H S - K - - - - - - - - - - - - Q I M H R D L K S N N L L L G R N - - - - - - - - - - - - - - - - W T I K I C D F G F A K E I T I Q N - - - - - - - - - - - - - - - - - - - - P L S M T I C G T D E F M S P E V I L G - - - - - - - - - - - - - - - - - - - - - - I Q Y S Y S A D I Y S F G M V L L E L I T R S - - - - - - - - - - - - - - - - K L D E R L P Q N - N F D - - - - - - - - - - - - - - - - - - - - - - - - - - - I - - - D Y E E L Q N K - - - - - - - - - I P S E C - - - - - P - - - R E F - - - L E L S M K C C N Y D P N D R P S F T D I V Q T - - - - - |
| LRRKb_Spur |
- - - - - - - - - - - - - N L L G D G S F G - S V Y K G T - Y R - - - - - D K - - - - - - P - V A V K V F S K G G E S - - - - - - - - - - - - - - - - - - - - - - - - - - - H P H R L L R Q E V T V - - L R - - H - - - - L - Q - H P S L V S M E G V G L N - - - - - - - - - - - - - P - - R I L V M E L A P - L G S - L N T - - - - - L L A S G - R - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - I K N R N L Q H R I A M Q V A E G L A F L H K - N - - - - - - - - - - - - R I V Y R D L K P D N V L I F S L A L G - - - - - - - - - - T - L Q N A K I S D Y G I S R F A T P Y G L - - - - - - - - - - - - - - - - - - - - - - K S S E G T P G Y R A P E V I R - - G - - - - - - - - - - - - - - - - - - - T S N Y N T E V D I Y S F G I L L Y E L V S E G H - - - - - - - - - - - - - - - R P F E E M E F R - T - - - E I - - - - - - - - - - - - - - - - - - - - - - - - D E A V I K G Q H L P P - I S Q G - - - - - G V A P - - - - - W - - - P D L - - - Q E L I D H C T E H V P E S R P T A Q E A F E K - - - - - |
| HH498_Mmus |
- - - - - - - - - - E F H E I I G S G S F G - K V Y K G R - C R - - - - - N K - - - - I - - - V A I K R Y R A N T Y C - - - - - - - - - - - - - - - - - - - - - - S K - - - S D V D M F C R E V S I - - L C - - Q - - - L - - N - H P C V V Q F V G A C L D D P - - - - - - - - - - - - S Q F A I V T Q Y I S - G G S - L F S - - - - - L L H E Q K - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - R I L D L Q S K L I I A V D V A K G M E Y L H S - L - - - - - - - - - - T Q P I I H R D L N S H N I L L Y E D - - - - - - - - - - - - - - - - G H A V V A D F G E S R F L Q S L D E - - - - - - - - - - - - - - - - - - - D N M T K Q P G N L R W M A P E V F T Q C - - - - - - - - - - - - - - - - - - - - - T R Y T I K A D V F S Y A L C L W E L L T G E - - - - - - - - - - - - - - - - I P F A H L K P A - A A A - - - - - - - - - - - - - - - - - - - - - - - - - A - - - D M A Y H H I R P P - - - - - - - - - I G Y S I - - - - - P - - - K P I - - - S S L L M R G W N A C P E G R P E F S E V V R K L E - - - |
| RAF1_Mmus |
- - - - - - - - - - - - S T R I G S G S F G - T V Y K G K - W H - - - - - G D - - - - - - - - V A V K I L K V V D P T - - - - - - - - - - - - - - - - - - - - - - - - - - P E Q L Q A F R N E V A V - - L R - - K - - - - T - R - H V N I L L F M G Y M T K D - - - - - - - - - - - - - - N L A I V T Q W C E - G S S - L Y K - - - - - H L H V Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E T K F Q M F Q L I D I A R Q T A Q G M D Y L H A - K - - - - - - - - - - - - N I I H R D M K S N N I F L H E - - - - - - - - - - - - - - - - G L T V K I G D F G L A T V K S R W S G S - - - - - - - - - - - - - - - - - - Q Q V E Q P T G S V L W M A P E V I R M Q D - - - - - - - - - - - - - - - - - - - D N P F S F Q S D V Y S Y G I V L Y E L M A G E - - - - - - - - - - - - - - - - L P Y A H I N N R - D Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - I I F M V G R G Y A S P D L S R L - - - - - Y K N C - - - - - P - - - K A M - - - K R L V A D C V K K V K E E R P L F P Q I L S S I - - - - |
| rk2_Ddis |
- - - - - - - - - - - I G V R I G K G N F G - E V Y L G T - W R - - - - - G S - - - - Q - - - V A V K K L P A H N I - - - - - - - - - - - - - - - - - - - - - - - - - - N E N I L K E F H R E I N L - - M K - - N - - - L - - R - H P N V I Q F L G S C L I S P - - - - - - - - - - - - - D I C I C T E Y M P - R G S - L Y S - - - - - I L H N E - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K I K I S W S L V K R M M I D A A K G I I Y L H G - S - - - - - - - - - T P - V I L H R D L K S H N L L V D E - - - - - - - - - - - - - - - - N W K V K V A D F G L S T I E Q Q - - - - - - - - - - - - - - - - - - - - - - G A T M T A C G T P C W T S P E V L R S - - - - - - - - - - - - - - - - - - - - - - Q R Y T E K A D V Y S F G I I L W E C A T R Q - - - - - - - - - - - - - - - - D P Y F G I P P F - Q V - - - - - - - - - - - - - - - - - - - - - - - - - - - - I F A V G R E G M R P P - - - - - - - - - T P K Y G - - - - - P - - - P K Y - - - I Q L L K D C L N E N P S Q R P T M E Q C L E I - - - - - |
| DDB0214883_Ddis |
- - - - - - - - - - - I G A R I G R G G Y G - Q V F R G S - W R - - - - - G T - - - - E - - - V A V K M L F N D N V - - - - - - - - - - - - - - - - - - - - - - - - - - N L K L I S D L R K E V D L - - L C - - K - - - L - - R - H P N I V L F M G A C T E P S - - - - - - - - - - - - - S P C I V T E Y L S - R G S - L A N - - - - - I L L D E - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - S I E M D W G L R L Q L G F D C A R G M T Y L H S - R - - - - - - - - - N P - I I I H R D L K T D N L L V D D - - - - - - - - - - - - - - - - S W Q V K V A D F G L A T V K S H - - - - - - - - - - - - - - - - - - - - - - T F A K T M C G T T G W V A P E V L A E - - - - - - - - - - - - - - - - - - - - - - E G Y T E K A D V Y S Y A I V L W E L L T R L - - - - - - - - - - - - - - - - I P Y A G K N T M - Q V - - - - - - - - - - - - - - - - - - - - - - - - - - - - V R S - I D R G E R L P - - - - - - - - - M P A W C - - - - - P - - - P K Y - - - A A L M N R C W E T D P T H R P S F P E I L P - - - - - - |
| ANKRD3_Mmus |
- - - - - - - - - - - - - - K V G S G G F G - Q V Y K V R H V H - - - - - W K - - - - - - T W L A I K C S P S L H V D D - - - - - - - - - - - - - - - - - - - - - - R E R M E - - - - L L E E A K K - - M E - - M - - - - A - K - F R Y I L P V Y G I C Q - - - - - - - - - - - - - E P - - V G L V M E Y M E - T G S - L E K - - - - - L L A S - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E P L P W D L R F R I V H E T A V G M N F L H C - M - - - - - - - - - S P - P L L H L D L K P A N I L L D - - - - - - - - - - - - - - - A - H Y H V K I S D F G L A K C N G M S H S - H - - - - - - - - - - - - - - - - D L S M D G L F G T I A Y L P P E R I R E K - - - S - - - - - - - - - - - - - - - - - R L F D T K H D V Y S F A I V I W G V L T Q K - - - - - - - - - - - - - - - - K P F A D E K N I - L H - - - - - - - - - - - - - - - - - - - - - - - - - - - - I M M K V V K G H R P E - L P P I C - R P R - P R A - - - - - C - - - A S L - - - I G L M Q R C W H A D P Q V R P T F Q E I T S E - - - - - |
| Raf_Spur |
- - - - - - - - - - - L G S R I G A G S F G - T V F S G Q - W H - - - - - G S - - - - - - - - V A V K R L N V K D P T - - - - - - - - - - - - - - - - - - - - - - - - - - P S Q L Q A F K N E V A V - - L R - - K - - - - T - R - H A N I L L F M G C T S K P - - - - - - - - - - - - - - Q L A I V T Q W C E - G S S - L Y K - - - - - H L H V L - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - D T K L V M H Q L I D I S R Q T A Q G M D Y L H A - K - - - - - - - - - - - - N I I H R D L K S N N I F L H D - - - - - - - - - - - - - - - - D L T V K I G D F G L A T V K S R W S G S - - - - - - - - - - - - - - - - - - Q Q F E Q P S G S I L W M A P E V I R M R D - - - - - - - - - - - - - - - - - - - T N P Y S F Q S D V Y A F G V V L F E L V T Q S - - - - - - - - - - - - - - - - L P Y Q N I K H K - D Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - I I W M V G R G Y L Q P D L S K V - - - - - R N D T - - - - - P - - - K A L - - - K R L I T D C Y A L N R D E R P L F P V V L A - - - - - - |
| ANKK5_Spur |
- - - - - - - - - - - - - - - - G G G S F G - E V F K A T H A V - - - - - H G - - - - - - D - V A V K L A K G - - - - Q - - - - - - - - - - - - - - - - - - - - V V M Y Q K E - - - - - - - A Q K H - - E N - - S - - - - L - I - C E H I V S I K G F V E C K D D - - - - - - A C T C G - L I G I V M T Y M E - N G S - L W D - - - - - F R T T K W L H - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - H P D L W P L T N R M I Y Q I S C G M H F L H S - N - - - - - - - - - - - - D I I H R D L K L E N V L V D - - - - - - - - - - - - - - - G - D L N V K I S D L G L A T N L Q T - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - |
| put_Dmel |
- - - - - - - - - - - - - - - - - - G R F G - D V W Q A K - L N - - - - - - - - - - N Q - D - V A V K I F R M Q E - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K E S W T T E H D I Y K L P - - R - - - - M - R - H P N I L E F L G V E K H M - - - - - - - - - D - - K P E Y W L I S T Y Q H - N G S - L C D - - - - - Y L K S H - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - T I S W P E L C R I A E S M A N G L A H L H E E I P A S - K T D G L K P - S I A H R D F K S K N V L L K S - - - - - - - - - - - - - - - - D L T A C I A D F G L A M I F Q P G K - - - - - - - - - - - - - - - - P C G - - D T H G Q V G T R R Y M A P E V L E G A I N F N - - - - - - - - - R - - - - - - - D - A F L R I D V Y A C G L V L W E M V S R C D - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - |
| IRAK4_Mmus |
- - - - - - - - - - - - - - R M G E G G F G - V V Y K G C - V N - - - - - N T - - - - - - I - V A V K K L G A M - - V E - - - - - - - - - - - - - - - - - - - - - - I S T E E L K Q Q F D Q E I K V - - M A - - T - - - - C - Q - H E N L V E L L G F S S D S - - - - - - - - - - - - - D N L C L V Y A Y M P - N G S - L L D - - - - - R L S C L D G - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - T P P L S W H T R C K V A Q G T A N G I R F L H E - N - - - - - - - - - - - - H H I H R D I K S A N I L L D - - - - - - - - - - - - - - - K - D F T A K I S D F G L A R A S A R L A - - - - - - - - - - - - - - - - - - Q T V M T S R I V G T T A Y M A P E A L R G - - - - - - - - - - - - - - - - - - - - - - - E I T P K S D I Y S F G V V L L E L I T G L A A V - - - - - - - D E N R E P Q L L L D I K E E - I E D E E K T - - - - - - - - - - - - - - - - - - - - - - - - - - - I E D Y T D E K - M S D A - - - - - D P A S - - - - - V - - - E A M - - - Y S A A S Q C L H E K K N R R P D I A K V Q Q - - - - - - |
| ALK2_Mmus |
- - - - - - - - - - - L L E C V G K G R Y G - E V W R G S - W Q - - - - - - - - - - G E - N - V A V K I F S S R D - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E K S W F R E T E L Y N T V - - M - - - - L - R - H E N I L G F I A S D M T S - - - - - - - - - R H S S T Q L W L I T H Y H E - M G S - L Y D - - - - - Y L Q L T - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - T L D T V S C L R I V L S I A S G L A H L H I E I F G T Q G - - - - K S - A I A H R D L K S K N I L V K K - - - - - - - - - - - - - - - - N G Q C C I A D L G L A V M H S Q S T - - - - - - - - - - - - - - - - N Q L D V G N N P R V G T K R Y M A P E V L D E T I Q V D - - - - - - - - - C - - - - - - F D - S Y K R V D I W A F G L V L W E V A R R M V S N - - - - - - G I V E D Y K P P F Y D V V P N - D P S F - - - - - - - - - - - - - - - - - - - - - - - E D M R K V V C V D Q Q R P N - I P N R - - W F - - S - D - - - - - - - - - P T L T S L A K L M K E C W Y Q N P S A R L T A L R I K K T L - - - - |
| DDB0229866_Ddis |
- - - - - - - - - - - - - - E I G K G A Y G - K I F K G E - Y F - - - - - G T - - - - P - - - V A I K E I S L Q P N - - - - - - - - - - - - - - - - - - - - - - - D V K Y K D L T K F I Q R E V A M - - L - - - R - - - F - - S - H P N L V Q F I G V S E R - - - - - - - - - - - - - G S S L Y I V T E F V Q - G G D - L A Y - - - - - Y L F R N K F D D T P E Q Y I H R K V N V G S S S T P D L S D P T V H N G E K L V Q L A W P L R I K I A Y D V A C A M A Y L H S - R - - - - - - - - - - - - N V I H R D L K S T N L L V G D N - - - - - - - - - - - - - - - - W R I K V C D M G F A R T A Q V G G G S - - - - - - - - - - - - - - - - R A K R T M T I C G T T N C M A P E V V L G - - - - - - - - - - - - - - - - - - - - - - Q D Y N E A C D V F S Y G I V L S E I I T R M - - - - - - - - - - - - - - - - D T T N N L R P S - S L K - - - - - - - - - - - - - - - - - - - - - - - - - - - Y G L D V - D V L L P L - - - - - - - - - V P K D C - - - - - P - - - P P F - - - L K L V L D C T E Y D P D N R P T F K E I T E R L K - - - |
| KSR2_Hsap |
- - - - - - - - - - E I G E L I G K G R F G - Q V Y H G R - W H - - - - - G E - - - - - - - - V A I R L I D I E R D - - - - - - - - - - - - - - - - - - - - - - - - - - N E D Q L K A F K R E V M A - - Y R - - Q - - - - T - R - H E N V V L F M G A C M S P - - - - - - - - - - - - - P H L A I I T S L C K - G R T - L Y S - - - - - V V R D A - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K I V L D V N K T R Q I A Q E I V K G M G Y L H A - K - - - - - - - - - - - - G I L H K D L K S K N V F Y D - - - - - - - - - - - - - - - - - N G K V V I T D F G L F S I S G V L Q A - - - - - - - - - - - - - - - G R R E D K L R I Q N G W L C H L A P E I I R Q L S P D T - - - - - - - - E - - - - - E D K L P F S K H S D V F A L G T I W Y E L H A R E - - - - - - - - - - - - - - - - W P F K T Q P A E - A I I - - - - - - - - - - - - - - - - - - - - - - - - - - - W Q M G - - T G M K P N - L S Q I G - - - - - - - M - - - - - G - - - K E I - - - S D I L L F C W A F E Q E E R P T F T K L M D - - - - - - |
| Rck1_Ddis |
- - - - - - - - - - - I H K W I A S G S S G - R V Y N G Q - Y K - - - - - G K - - - - D - - - V A I K V L G P E V C - - - - - - - - - - - - - - - - - - - - - - - - - - V H F D L N E F K R E V A L - - M S - - I - - - F - - K - H D N L A R C L G A G Q Y D D - - - - - - - - - - - - - K Y F H L T E Y C H - N G S - L F S - - - - - Y L R D Q R - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - N N I S F G Q R L H F A L G I A R G M R Y L H S - M - - - - - - - - - - - - S I I H R D L K S M N I L L T K R L - - - - - - - - - - - - - - - - K I K I V D F G T S R V A N K Y - - - - - - - - - - - - - - - - - - - - - - N M T T H V G T Q A W M A P E I F T S - - - - - - - - - - - - - - - - - - - - - - R T Y T N K V D V Y S Y A I I L F E I F T R K - - - - - - - - - - - - - - - - S A Y D E N A N I - N I P - - - - - - - - - - - - - - - - - - - - - - - - - - - - - N M V M K G E R P E - - - - - - - - - L P K D M - - - - - Q - - - T S I - - - S N I I K K C W Q Q K P S N R P S F I K I V A Y L - - - - |
| MLK_Spur |
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K I T D F G L A R E M Y K - - - - - - - - - - - - - - - - - - - - - - T T R M S A A G T Y A W M A P E V I K S - - - - - - - - - - - - - - - - - - - - - - S L F S K S S D V W S F G I L L W E L L T G E - - - - - - - - - - - - - - - - V P Y K G I D T L - A V A - - - - - - - - - - - - - - - - - - - - - - - - - Y - - - G I A V N K L T L P - - - - - - - - - I P S T C - - - - - P - - - E I F - - - S K M L L D C W N Y D P H E R P T F S E I M Q Q L - - - - |
| Shk5_Ddis |
- - - - - - - - - - - - - - L L G G G A Y G - K V Y K A T - C R - - - - - G K - - - - K - - - V A V K V P K K Q T L - - - - - - - - - - - - - - - - - - - - - - - - - - S E S E L K S F K N E V E I - - M K - - Q - - - I F - - - H P N V V L C L G A C T K P G - - - - - - - - - - - - - K V M I V S E L M Q - - T D - L E K - - - - - L I H S S E - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - V E P P P L Y E R M K M C L D A A L G I N W L H G - I - - - - - - - - - C N - - I I H R D L K L A N L M I S K - - - - - - - - - - - - - - - - D K T V K I G D F G F S Q V I K T G T T - - - - - - - - - - - - - - - - - - L S D Q K G P K G T A L Y M A P E V M M K - - - - - - - - - - - - - - - - - - - - - - H E F N E K A D V Y S F G L I L Y E M A T C E - - - - - - - - - - - - - - - - E L F P E Y S E - - I - - - - - - - - - - - - - - - - - - - - - - - - - - D P F Y D A I C N K K L R P P - - - - - - - - - I P D S F - - - - - P - - - K S L - - - K T L I Q K C W D H D P N K R P S F N E V T Q R M - - - - |
| TAK1_Hsap |
- - - - - - - - - - - V E E V V G R G A F G - V V C K A K - W R - - - - - A K - - - - D - - - V A I K Q I E S E S E - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - R K A F I V E L R Q - - L S - - R - - - V - - N - H P N I V K L Y G A C L N - - - - - - - - - - - - - - - P V C L V M E Y A E - G G S - L Y N - - - - - V L H G A - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E P L P Y Y T A A H A M S W C L Q C S Q G V A Y L H S - M - - - - - - - - - Q P K A L I H R D L K P P N L L L V A G - - - - - - - - - - - - - - - G T V L K I C D F G T A C D I Q T - - - - - - - - - - - - - - - - - - - - - - - H M T N N K G S A A W M A P E V F E G - - - - - - - - - - - - - - - - - - - - - - S N Y S E K C D V F S W G I I L W E V I T R R - - - - - - - - - - - - - - - - K P F D E I G G P - A F R - - - - - - - - - - - - - - - - - - - - - - - - - - - I M W A V H N G T R P P - - - - - - - - - L I K N L - - - - - P - - - K P I - - - E S L M T R C W S K D P S Q R P S M E E I V K I - - - - - |
| TESK1_Hsap |
- - - - - - - - - - - - A E K I G A G F F S - E V Y K V R - H R - - - - - Q S - - - G Q - - - V M V L K M N K - - - - - - - - - - - - - - - - - - - - - - - - - - - - - L P S N R G N T L R E V Q L - - M N - - R - - - L - - R - H P N I L R F M G V C V H Q G - - - - - - - - - - - - - Q L H A L T E Y M N - G G T - L E Q - - - - - L L S S P - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E P L S W P V R L H L A L D I A R G L R Y L H S - K - - - - - - - - - G - - - V F H R D L T S K N C L V R R E - D - R - - - - - - - - - - - G F T A V V G D F G L A E K I P V Y R - - - - - - - - - - - - - - - - E G A R K E P L A V V G S P Y W M A P E V L R G - - - - - - - - - - - - - - - - - - - - - - E L Y D E K A D V F A F G I V L C E L I A R V - - - - - - - - - - - - - - - - P A D P D Y L P R - T E D - - - - - - - - - - - - - - - - - - - - - - - - F G - - - L D V P A F R T - L - - - - - - - - - V G D D C - - - - - P - - - L P F - - - L L L A I H C C N L E P S T R A P F T E I T Q H L E - - - |
| MLK1_Mmus |
- - - - - - - - - - - - E E I I G I G G F G - K V Y R A F - W A - - - - - G D - - - - E - - - V A V K A A R H D P D E - - - - - - - - - - - - - - - - - - - - - - D I - - S Q T I E N V R Q E A K L - - F A - - M - - - L - - K - H P N I I A L R G V C L K E - - - - - - - - - - - - - P N L C L V M E F A R - G G P - L N R - - - - - V L S G - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K R I P P D I L V N W A V Q I A R G M N Y L H D - E - - - - - - - - - A I V P I I H R D L K S S N I L I L Q K V E N G - - - - - - - D L S - N K I L K I T D F G L A R E W H R - - - - - - - - - - - - - - - - - - - - - - T T K M S A A G T Y A W M A P E V I R A - - - - - - - - - - - - - - - - - - - - - - S M F S K G S D V W S Y G V L L W E L L T G E - - - - - - - - - - - - - - - - V P F R G I D G L - A V A - - - - - - - - - - - - - - - - - - - - - - - - - Y - - - G V A M N K L A L P - - - - - - - - - I P S T C - - - - - P - - - E P F - - - A K L M E D C W N P D P H S R P S F T S I L D Q - - - - - |
| IRAK3_Hsap |
- - - - - - - - - - - K D F L I G E G E I F - E V Y R V E - I Q - - - - - N L - - - - - - T - Y A V K L F K Q E - - K K - - - - - - - - - - - - - - - - - - - - - - M Q C K K H W K R F L S E L E V - - L L - - L - - - - F - H - H P N I L E L A A Y F T E T - - - - - - - - - - - - - E K F C L I Y P Y M R - N G T - L F D - - - - - R L Q C V G D - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - T A P L P W H I R I G I L I G I S K A I H Y L H N - V - - - - - - - Q - P C - S V I C G S I S S A N I L L D - - - - - - - - - - - - - - - D - Q F Q P K L T D F A M A H F R S H L E - - - - - - - - - - - - - - H Q S C T I N M T S S S S K H L W Y M P E E Y I R Q - - - - - - - - - - - - - - - - - - - - - - G K L S I K T D V Y S F G I V I M E V L T G C R V V - - - - - - - L D D P K H I Q L R D L L R E - L - M E K R - - - - - - - - - - - - - - - - - - - - - - - - G L D S C L S F L D K K - V P P C - - - - - P R N F - - - - - S - - - A K L - - - F C L A G R C A A T R A K L R P S M D E V L N T - - - - - |
| Roco8_Ddis |
- - - - - - - - - - - - Q E E I G V G G F S - R V Y K - G I Y K - - - - - N N - - - - - - T - V A I K Q F N F E R M D L - - - - - - - - - - - - - - - - - - - - I - - - - - - D S T S F N N - L N S - - L T - - - - - - - - - - - - - - - - - - - - - I S - - - - - - - - - - - - - P S - N S S L S I S L S S - S T S S L S P - - - - - P I V N N N - - - - - - - - - - - - - - - - N N N N N L N N N L N N L N N N N K L Y I Q Q Q Q Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - |
| ACTR2B_Hsap |
- - - - - - - - - - - - - - - - - - G R F G - C V W K A Q - L M - - - - - - - - - - N D - F - V A V K I F P L Q D - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K Q S W Q S E R E I F S T P - - G - - - - M - K - H E N L L Q F I A A E K R G - - - - - - - - - S N L E V E L W L I T A F H D - K G S - L T D - - - - - Y L K G N - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - I I T W N E L C H V A E T M S R G L S Y L H E D V P W C R G - E G H K P - S I A H R D F K S K N V L L K S - - - - - - - - - - - - - - - - D L T A V L A D F G L A V R F E P G K - - - - - - - - - - - - - - - - P P G - - D T H G Q V G T R R Y M A P E V L E G A I N F Q - - - - - - - - - R - - - - - - - D - A F L R I D M Y A M G L V L W E L V S R C K A A - - - - - D G P V D E Y M L P F E E E I G Q - H P S L - - - - - - - - - - - - - - - - - - - - - - - E E L Q E V V V H K K M R P T - I K D H - - W L - - K - H - - - - - - - - - P G L A Q L C V T I E E C W D H D A E A R L S A G - - - - - - - - - - |
| DDB0229850_Ddis |
- - - - - - - - - - - - - C R L G S G S L C - R V Y K G R - L N - - - - - G K - - - - - - P - V A I K V F S P I - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - R F E E F K T E F L M - - M Q - - S - - - - L - R S S P F L I S F Y G V S I V E E - - - - - - - - - P Q K Q C Y C I I T E F C S - R D S - L Y H - - - - - I M T D R L I - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E I G W N R F F Q F S M Q I I L G L Q S L H N - R - - - - - - - - K P K - P I V H R D V T S L N I L V N - - - - - - - - - - - - - - - E - D W E C K I S N F S A S R F N C L N T E Y I - - - - - - - - - - - - - - - - - - N S N N Q N K S F A F C S P E S S D F Q D I D D - - - - - - - - - - - D Y T S L S S S I T S K S D I Y S F G I I M F E L I S R I I N G - - - - - - - - - - E Y S H P F S E F K D I - K - - - N D - - - - - - F - - - - - - - - - - - - - Q L L - - - L S S K N G L R P S - L P N I - - - - - - - - C - - - - - P - - - E P L - - - E K L Y K Q C V D Q S P L N R P S C E E V I I - - - - - - |
| ACTR2B_Mmus |
- - - - - - - - - - - - - - - - - - G R F G - C V W K A Q - L M - - - - - - - - - - N D - F - V A V K I F P L Q D - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K Q S W Q S E R E I F S T P - - G - - - - M - K - H E N L L Q F I A A E K R G - - - - - - - - - S N L E V E L W L I T A F H D - K G S - L T D - - - - - Y L K G N - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - I I T W N E L C H V A E T M S R G L S Y L H E D V P W C R G - E G H K P - S I A H R D F K S K N V L L K S - - - - - - - - - - - - - - - - D L T A V L A D F G L A V R F E P G K - - - - - - - - - - - - - - - - P P G - - D T H G Q V G T R R Y M A P E V L E G A I N F Q - - - - - - - - - R - - - - - - - D - A F L R I D M Y A M G L V L W E L V S R C K A A - - - - - D G P V D E Y M L P F E E E I G Q - H P S L - - - - - - - - - - - - - - - - - - - - - - - E E L Q E V V V H K K M R P T - I K D H - - W L - - K - H - - - - - - - - - P G L A Q L C V T I E E C W D H D A E A R L S A G - - - - - - - - - - |
| Shk1_Ddis |
- - - - - - - - - - - T E S I L G D G S F G - T V Y K G R - C R - - - - - L K - - - - D - - - V A V K V M L K Q V - - - - - - - - - - - - - - - - - - - - - - - - - - - D Q K T L T D F R K E V A I - - M S - - K - - - I F - - - H P N I V L F L G A C T S T P - - - - - - - - - - - - G K L M I C T E L M - - K G N - L E S - - - - - L L L D P - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - M V K L P L I T R M R M A K D A A L G V L W L H S - S - - - - - - - - - N P - V F I H R D L K T S N L L V D A - - - - - - - - - - - - - - - - N L T V K V C D F G L S Q I K Q R G E N - - - - - - - - - - - - - - - - - L K D G Q D G A K G T P L W M A P E V L Q G - - - - - - - - - - - - - - - - - - - - - - R L F N E K A D V Y S F G L V L W Q I F T R Q - - - - - - - - - - - - - - - - E L F P E F D N - - F - - - - - - - - - - - - - - - - - - - - - - - - - - F K F V A A I C E K Q L R P S - - - - - - - - - I P D D C - - - - - P - - - K S L - - - K E L I Q K C W D P N P E V R P S F E G I V S E L E - - - |
| LRRK1_Mmus |
- - - - - - - - - - - - - - I L G Q G G S G T V I Y Q A R - Y Q - - - - - G Q - - - - - - P - V A V K R F H I K K F K N S A N A P A D T M - - - - - - - L R H L R A M D A M K N F S D F R Q E A S M - - L H - - A - - - - L - Q - H P C I V S L I G I S I H - - - - - - - - - - - - - P - - L C F A L E L A P - L G S - L N T - - - - - V L S E N A K - - - - - - - - - - - - - - - - - - - - - - - - - - - - - D S S F M P L G H M L T Q K I A Y Q I A S G L A Y L H K - K - - - - - - - - - - - - N I I F C D L K S D N I L V W S L S A K - - - - - - - - - - E - H I N I K L S D Y G I S R Q S F H E G A - - - - - - - - - - - - - - - - - - - - - - L G V E G T P G Y Q A P E I R P - - - - - - - - - - - - - - - - - - - - - - R I V Y D E K V D M F S Y G M V L Y E L L S G Q - - - - - - - - - - - - - - - - R P A L G H H Q L - Q - - - - I - - - - - - - - - - - - - - - - - - - - - - - - V K K L S K G - I R P V - L G Q P - - - - - E E V Q - - - - - F - - - H R L - - - Q A L M M E C W D T K P E K R P L A L S V V S Q - - - - - |
| IRAK4_Hsap |
- - - - - - - - - - - - - - K M G E G G F G - V V Y K G Y - V N - - - - - N T - - - - - - T - V A V K K L A A M - - V D - - - - - - - - - - - - - - - - - - - - - - I T T E E L K Q Q F D Q E I K V - - M A - - K - - - - C - Q - H E N L V E L L G F S S D G - - - - - - - - - - - - - D D L C L V Y V Y M P - N G S - L L D - - - - - R L S C L D G - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - T P P L S W H M R C K I A Q G A A N G I N F L H E - N - - - - - - - - - - - - H H I H R D I K S A N I L L D - - - - - - - - - - - - - - - E - A F T A K I S D F G L A R A S E K F A - - - - - - - - - - - - - - - - - - Q T V M T S R I V G T T A Y M A P E A L R G - - - - - - - - - - - - - - - - - - - - - - - E I T P K S D I Y S F G V V L L E I I T G L P A V - - - - - - - D E H R E P Q L L L D I K E E - I E D E E K T - - - - - - - - - - - - - - - - - - - - - - - - - - - I E D Y I D K K - M N D A - - - - - D S T S - - - - - V - - - E A M - - - Y S V A S Q C L H E K K N K R P D I K K V Q Q L L - - - - |
| ALK7_Mmus |
- - - - - - - - - - - L Q E I V G K G R F G - E V W H G R - W C - - - - - - - - - - G E - D - V A V K I F S S R D - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E R S W F R E A E I Y Q T V - - M - - - - L - R - H E N I L G F I A A D N K D - - - - - - - - - N G T W T Q L W L V S E Y H E - Q G S - L Y D - - - - - Y L N R N - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - I V T V A G M V K L A L S I A S G L A H L H M E I V G T Q G - - - - K P - A I A H R D I K S K N I L V K K - - - - - - - - - - - - - - - - C D T C A I A D L G L A V K H D S I M - - - - - - - - - - - - - - - - N T I D I P Q N P K V G T K R Y M A P E M L D D T M N L S - - - - - - - - - I - - - - - - F E - S F K R A D I Y S V G L V Y W E I A R R C S V G - - - - - - G V V E E Y Q L P Y Y D M V P S - D P S I - - - - - - - - - - - - - - - - - - - - - - - E E M R K V V C D Q K L R P N - L P N Q - - W Q - - S - C - - - - - - - - - E A L R V M G R I M R E C W Y A N G A A R L T A L R V K - - - - - - - |
| ALK2_Hsap |
- - - - - - - - - - - - - E C V G K G R Y G - E V W R G S - W Q - - - - - - - - - - G E - N - V A V K I F S S R D - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E K S W F R E T E L Y N T V - - M - - - - L - R - H E N I L G F I A S D M T S - - - - - - - - - R H S S T Q L W L I T H Y H E - M G S - L Y D - - - - - Y L Q L T - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - T L D T V S C L R I V L S I A S G L A H L H I E I F G T Q G - - - - K P - A I A H R D L K S K N I L V K K - - - - - - - - - - - - - - - - N G Q C C I A D L G L A V M H S Q S T - - - - - - - - - - - - - - - - N Q L D V G N N P R V G T K R Y M A P E V L D E T I Q V D - - - - - - - - - C - - - - - - F D - S Y K R V D I W A F G L V L W E V A R R M V S N - - - - - - G I V E D Y K P P F Y D V V P N - D P S F - - - - - - - - - - - - - - - - - - - - - - - E D M R K V V C V D Q Q R P N - I P N R - - W F - - S - D - - - - - - - - - P T L T S L A K L M K E C W Y Q N P S A R L T A L R I K K T - - - - - |
| KSR2_Mmus |
- - - - - - - - - - E I G E L I G K G R F G - Q V Y H G R - W H - - - - - G E - - - - - - - - V A I R L I D I E R D - - - - - - - - - - - - - - - - - - - - - - - - - - N E D Q L K A F K R E V M A - - Y R - - Q - - - - T - R - H E N V V L F M G A C M S P - - - - - - - - - - - - - P H L A I I T S L C K - G R T - L Y S - - - - - V V R D A - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - K I V L D V N K T R Q I A Q E I V K G M G Y L H A - K - - - - - - - - - - - - G I L H K D L K S K N V F Y D - - - - - - - - - - - - - - - - - N G K V V I T D F G L F S I S G V L Q A - - - - - - - - - - - - - - - G R R D D K L R I Q N G W L C H L A P E I I R Q L S P D T - - - - - - - - E - - - - - E D K L P F S K H S D V F A L G T I W Y E L H A R E - - - - - - - - - - - - - - - - W P F K T Q P A E - A I I - - - - - - - - - - - - - - - - - - - - - - - - - - - W Q M G - - T G M K P N - L S Q I G - - - - - - - M - - - - - G - - - K E I - - - S D I L L F C W A F E Q E E R P T F T K L M D - - - - - - |
| TGFBR1_Mmus |
- - - - - - - - - - - - - E S I G K G R F G - E V W R G K - W R - - - - - - - - - - G E - E - V A V K I F S S R E - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E R S W F R E A E I Y Q T V - - M - - - - L - R - H E N I L G F I A A D N K D - - - - - - - - - N G T W T Q L W L V S D Y H E - H G S - L F D - - - - - Y L N R Y - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - T V T V E G M I K L A L S T A S G L A H L H M E I V G T Q G - - - - K P - A I A H R D L K S K N I L V K K - - - - - - - - - - - - - - - - N G T C C I A D L G L A V R H D S A T - - - - - - - - - - - - - - - - D T I D I A P N H R V G T K R Y M A P E V L D D S I N M K - - - - - - - - - H - - - - - - F E - S F K R A D I Y A M G L V F W E I A R R C S I G - - - - - - G I H E D Y Q L P Y Y D L V P S - D P S V - - - - - - - - - - - - - - - - - - - - - - - E E M R K V V C E Q K L R P N - I P N R - - W Q - - S - C - - - - - - - - - E A L R V M A K I M R E C W Y A N G A A R L T A L R I K K - - - - - - |
| TAK1_Mmus |
- - - - - - - - - - - V E E V V G R G A F G - V V C K A K - W R - - - - - A K - - - - D - - - V A I K Q I E S E S E - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - R K A F I V E L R Q - - L S - - R - - - V - - N - H P N I V K L Y G A C L N - - - - - - - - - - - - - - - P V C L V M E Y A E - G G S - L Y N - - - - - V L H G A - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E P L P Y Y T A A H A M S W C L Q C S Q G V A Y L H S - M - - - - - - - - - Q P K A L I H R D L K P P N L L L V A G - - - - - - - - - - - - - - - G T V L K I C D F G T A C D I Q T - - - - - - - - - - - - - - - - - - - - - - - H M T N N K G S A A W M A P E V F E G - - - - - - - - - - - - - - - - - - - - - - S N Y S E K C D V F S W G I I L W E V I T R R - - - - - - - - - - - - - - - - K P F D E I G G P - A F R - - - - - - - - - - - - - - - - - - - - - - - - - - - I M W A V H N G T R P P - - - - - - - - - L I K N L - - - - - P - - - K P I - - - E S L M T R C W S K D P S Q R P S M E E I V K I - - - - - |
| 6778_Tthe |
- - - - - - - - - - K M G N K I G S G N S C - E V Y K G R - C K - - - - - N K - - - - D - - - V A I K V I K L D N S - - - - - - - - - - - - - - - - - - - - - - - - - - D E L H L K E F Q R E I Q A - - L I - - K - - - L K - N - H E N L L Q L V A I S S Y D N - - - - - - - - - - - - - N F Y I I T E F C E - G G T - L F E - - - - - L L H K K K - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - Q - Q I K M I K W D H R I R W L K D L A K G M L Y L H Q - N - - - - - - - - - - - - N L I H R D L K S L N L L F D K P Y D P N - - - - - - - S D N - I P T M K I A D F G L T R V G S S Q A V - - - - - - - - - - - - - - - - - - - P F Q S G A L G T F H W M A P E T M R N - - - - - - - - - - - - - - - - - - - - - - Q P V S T Q A D V Y S F A I V M Y E L A S R E - - - - - - - - - - - - - - - - T P F K E K G N V - A A I - - - - - - - - - - - - - - - - - - - - - - - - - - - - - I K Y V S E E K G R - P N L Q K - - - L H F D T - - - - - P - - - Q F I - - - I K L M I K C W D E N P K K R P K F D Q I I Q V L - - - - |
| LIMK1_Dmel |
- - - - - - - - - - - I G E K L G E G F F G - K V F K V T - H R - - - - - Q S - - - G E - - - V M V L K E L H R - A - - - - - - - - - - - - - - - - - - - - - - - - - - D E E A Q R N F I K E V A V - - L R - - L - - - L - - D - H R H V L K F I G V L Y K D K - - - - - - - - - - - - - K L H M V T E Y V A - G G C - L K E - - - - - L I H D P - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - A Q V L P W P Q R V R L A R D I A C G M S Y L H S - M - - - - - - - - - N - - - I I H R D L N S M N C L V R E D - R - - - - - - - - - - - - - - - S V I V A D F G L A R S V D - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - |
| TGFbR1_Hsap |
- - - - - - - - - - - - - E S I G K G R F G - E V W R G K - W R - - - - - - - - - - G E - E - V A V K I F S S R E - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - E R S W F R E A E I Y Q T V - - M - - - - L - R - H E N I L G F I A A D N K D - - - - - - - - - N G T W T Q L W L V S D Y H E - H G S - L F D - - - - - Y L N R Y - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - T V T V E G M I K L A L S T A S G L A H L H M E I V G T Q G - - - - K P - A I A H R D L K S K N I L V K K - - - - - - - - - - - - - - - - N G T C C I A D L G L A V R H D S A T - - - - - - - - - - - - - - - - D T I D I A P N H R V G T K R Y M A P E V L D D S I N M K - - - - - - - - - H - - - - - - F E - S F K R A D I Y A M G L V F W E I A R R C S I G - - - - - - G I H E D Y Q L P Y Y D L V P S - D P S V - - - - - - - - - - - - - - - - - - - - - - - E E M R K V V C E Q K L R P N - I P N R - - W Q - - S - C - - - - - - - - - E A L R V M A K I M R E C W Y A N G A A R L T A L R I K K - - - - - - |
| LZK_Mmus |
- - - - - - - - - - - - - - W L G S G A Q G - A V F L G K - F R - - - - - A E - - - - E - - - V A I K K V R E Q N E T - - - - - - - - - - - - - - - - - - - - - - D - - - - - - - - - - - - - I K H - - L R - - K - - - L - - K - H P N I I A F K G V C T Q A - - - - - - - - - - - - - P C Y C I I M E Y C A - H G Q - L Y E - - - - - V L R A - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - G R K I T P R L L V D W S T G I A S G M N Y L H L - H - - - - - - - - - - - - K I I H R D L K S P N V L V T H - - - - - - - - - - - - - - - - T D A V K I S D F G T S K E L S D - - K - - - - - - - - - - - - - - - - - - - S T K M S F A G T V A W M A P E V I R N - - - - - - - - - - - - - - - - - - - - - - E P V S E K V D I W S F G V V L W E L L T G E - - - - - - - - - - - - - - - - I P Y K D V D S S - A I I - - - - - - - - - - - - - - - - - - - - - - - - - W - - - G V G S N S L H L P - - - - - - - - - V P S T C - - - - - P - - - D G F - - - K I L M K Q T W Q S K P R N R P S F R Q T L M H L - - - - |
| DDB0229853_Ddis |
- - - - - - - - - - - - - - - I G R G S F G - Q V Q K A S - Y F - - - - - G T - - - - D - - - V A V K Q L S T L V S - - - - - - - - - - - - - - - - - - - - - - - - - I D P D Y F K F M L R E I K I - - L K - - G - - - M - - R - H P N I V Q Y I G A C C H E - - - - - - - - - - - - - G R Y M I V T E Y I K - G G D - L H Q - - - - - F I K A R G - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - V S N I S W T L R M K L A L D I A S A F S Y L H S - K - - - - - - - - - - - - K V I F R D L K A K N I L V D E I G D G - - - - - - - - - - - - L Y R A K V I D F G F A R I F D G K D - - - - - - - - - - - - - - - - - - - - T N N L T I C G S E N T M S P E V I V G - - - - - - - - - - - - - - - - - - - - - - S S Y N D S C D V Y S Y G V L L L E L I C G S - - - - - - - - - - - - - - - - R V V K T Q L K R - T P M - - - - - - - - - - - - - - - - - - - - - - - - - N A F D M - N L E K A E H L - - - - - - - - - A P E S C - - - - - P - - - R A F - - - M D L A K W C C S Y N P K D R P T F K - - - - - - - - - - |
| LIMK2_Mmus |
- - - - - - - - - - - - G E V L G K G F F G - Q A I K V T - H K - - - - - A T - - - G K - - - V M V M K E L I R - C - - - - - - - - - - - - - - - - - - - - - - - - - - D E E T Q K T F L T E V K V - - M R - - S - - - L - - D - H P N V L K F I G V L Y K D K - - - - - - - - - - - - - K L N L L T E Y I E - G G T - L K D - - - - - F L R S V - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - D - P F P W Q Q K V R F A K G I S S G M A Y L H S - M - - - - - - - - - C - - - I I H R D L N S H N C L I K L D - K - - - - - - - - - - - - - - - T V V V A D F G L S R L I V E E R K R P P V E K A T T K K R T L R K S D R K K R Y T V V G N P Y W M A P E M L N G - - - - - - - - - - - - - - - - - - - - - - K S Y D E T V D V F S F G I V L C E I I G Q V - - - - - - - - - - - - - - - - Y A D P D C L P R - T L D - - - - - - - - - - - - - - - - - - - - - - - - F G - - - L N V K L F W E K F - - - - - - - - - V P T D C - - - - - P - - - P A F - - - F P L A A I C C K L E P E S R P A F S K L E D S - - - - - |
| IRAK2_Hsap |
- - - - - - - - - - N Q N R K I S Q G T F A - D V Y R G H - R H - - - - - G K - - - - - - P - F V F K K L R E T - - A C - - - - - - - - - - - - - - - - - - - - - - S S P G S I E R F F Q A E L Q I - - C L - - R - - - - C - C - H P N V L P V L G F C A A R - - - - - - - - - - - - - Q F H S F I Y P Y M A - N G S - L Q D - - - - - R L Q G Q G G - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - S D P L P W P Q R V S I C S G L L C A V E Y L H G - L - - - - - - - - - - - - E I I H S N V K S S N V L L D - - - - - - - - - - - - - - - Q - N L T P K L A H P M A H L C P V N K - - - - - - - - - - - - - - - R S K Y T M M K T H L L R T S A A Y L P E D F I R V - - - - - - - - - - - - - - - - - - - - - - G Q L T K R V D I F S C G I V L A E V L T G I P A M - - - - - - - D N N R - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - |
| TKL1_Spur |
- - - - - - - - - - - - - - - V G K G Q F G - V V H R A S - W R - - - - - G T - - - - - - P - V A V K K I S - - I M G S - - - - - - - - - - - - - - - - - - - - - - - - - - - R K D E I E K E V A I - - H R - - R - - - - A - C - H P N I V Q M M A L G Y Q E - - - - - - - - - - - K - - E A F L V M Q F I E - G P S - L H T - - - - - V I F P D R D - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - D V K I P L D W A K K L Q I S R E V L Q A V T F M H A - C - - - - - - - - - - - - N I L H L D I K P A N I L V D S A T - - - - - - - - - - - - - - - I R P Y I C D L G L A H I K N R N L M - S - - - - - - - - - - - - - - - Q - - S A V N I R G T P C F M P P E A L G A T E - - - - - - - - - - - - - - - - - - P G Y R H S N K H D I W S V A G T L V E L Y S N K - - - - - - - - - - - - - - - - R L W G D S M N P - L A - - - - - - - - - - - - - - - - - - - - - - - - - - - - L M A R I L T G Q M S K - P E S L - - - - - - T A V - - - - - D - - - P K V - - - Q K I L E P C F R Q K P E K R P S A S D L L N Q - - - - - |
| Irak2_Spur |
- - - - - - - - - - - - A R R L G S G A F G - E V F H A Q - L D - - - - - M I - - - - - - D - V A I K K I I K N E N V K - - - - - - - - - - - - - - - - - - - - - - F N E S M I R G D Q I K E I E A - - L T - - K - - - - Y - R - H K N I V S L T A Y S F G E - - - - - - - - - - - - - D C I C L I F D F M K - N G S - L E Y - - - - - Q L E G R A - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - P Q G P L E W H R R H M I A E G A A L G I Q Y L H T - A - - - - - - - D S A V - P L I H Q D I K S A N I L L D - - - - - - - - - - - - - - - E - N F V P K I A D F G L A R P G P T Q G - - - - - - - - - - - - - - K E - Y T T I K T S V V F G T K A Y L A P E F M R D P - - - - - - - - - - - - - - - - - - - - K R K L S K K L D V Y S F G V V L L E L V T G E - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - |
| DDB0220436_Ddis |
- - - - - - - - - - - - - - - - G E G G Q C - S I Y K - - - - - - - - - - Y M - - - - - - G - T A M K R F K P S L S S S - - - - - - - - - - - - - - - - - - - - L - - - - - - I S K E F E N E V L I - - L E - - R - - - - L - N - H P N I V K I I T Y S T - - - - - - - - - - - - - I E - - R I I L L E F I D - G N S - L D K - - - - - Y P S Q S L P - - - - - - - - - - - - - - - - - - - - - - - - - - - - L S P S S S L P N P L K V I Q D F Q Q I V D A M I Y L H N - E - - - - - - - - - I - - G I I H F D L K P S N I L K N - - - - - - - - - - - - - - - S K N N K L K L I D F G I S K F L N N N N Q N N - - - - - - - - - - - - - - - - - N N N S L N M G S Y R Y S P P E L L C N N N Q N N - - - - - - - - - - - - - - - - - N L I N K S V D V F S F G I M L W E C L N W T - - - - - - - - - - - - - - - - L P Y E S L S R E - - Q - - - - - - - - - - - - - - - - - - - - - - - - - - - - V K Q I K T D M E R E S H L P L D H - L - - - P - - - - - - - - - - - K G I - - - Q D L I R L C W K H D P S I R P S F I E I R - - - - - - - |
| TGFbRII_Spur |
- - - - - - - - - - - L D E L I G K G R F G - A V W R A H - W K I N N G T - - - - K E R - T - V A V K V F L E Y D - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - S A S W N V E K E L F T D H S V M - - - - M - R - H E H I V Q F I S A E V R R V - - - - - - S V T S P T R Q Y W L I T K Y Y K - N G S - L H D - - - - - Y L R Q R - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - S V N W I D L C K L C G S A A R G L A H L H A E T Y G S H G - - L H K V - P V A H R D L K S T N I L V K D - - - - - - - - - - - - - - - - D G S C A I A D F G L A I R L D P Q S - - - - - - - - - - - - - - - - S T D H L A N S G Q V G T P R Y M S P E A L E C K V N L Q - - - - - - - - - D - - - - - - I E - S F K Q M D T Y A L A L I L W E I T S R C T L L - - - - - - K D I N E Y E L P F S R S L G D R R A S L - - - - - - - - - - - - - - - - - - - - - - - E M M K T L V V L R R E R P P - I T D E - - W L - - R - N - - - - - - - - - P G L T I I K N T V V D C W D E D P E A R L T - - - - - - - - - - - - |