About pdCSM-PPI
Running predictions
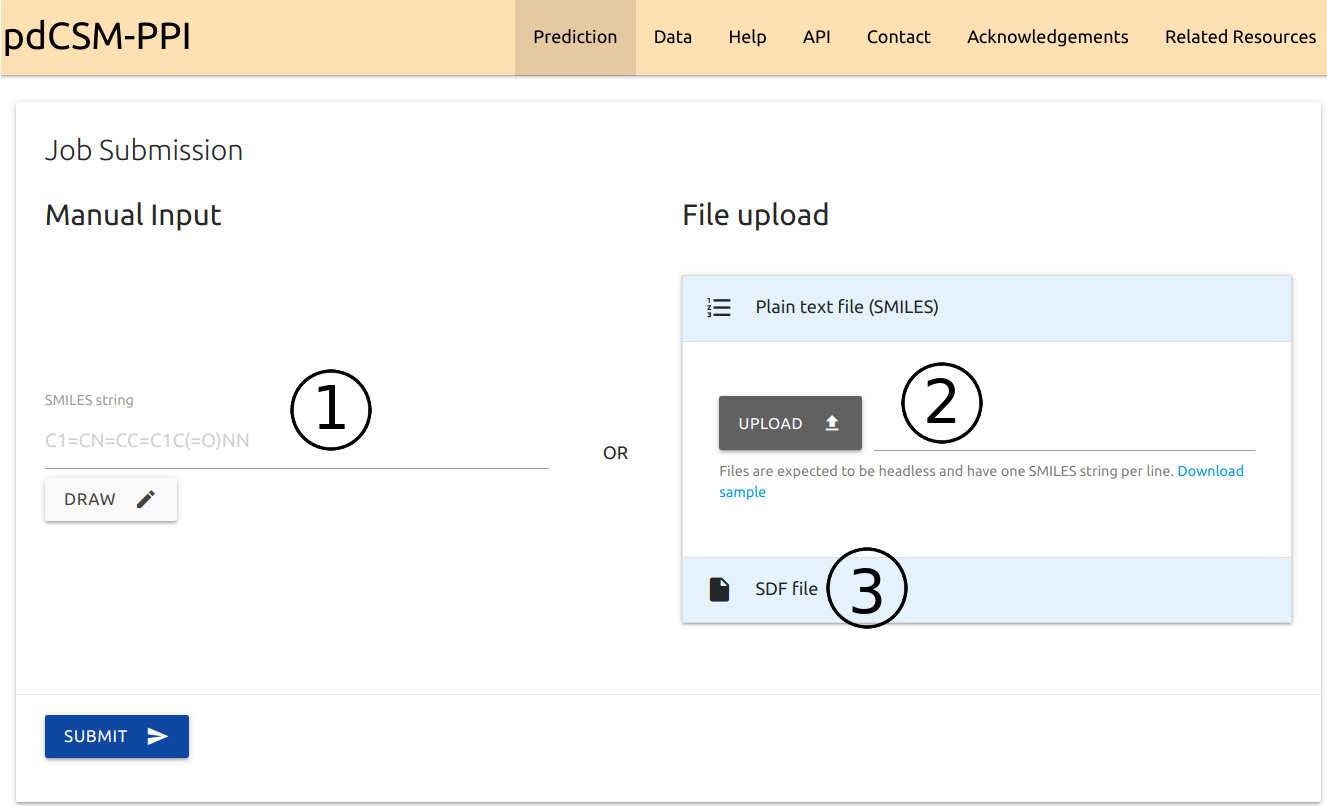
Input

- pdCSM-PPI can be queried with a single small-molecule in SMILES format (1). Users may also use the Kekule.js editor to draw the molecule by clicking the Draw button.
- Submit multiple compounds for batch processing by providing a headless plain text file with a list of SMILES, one per line (2) or
- as aStructure-Data File (SDF) (3).
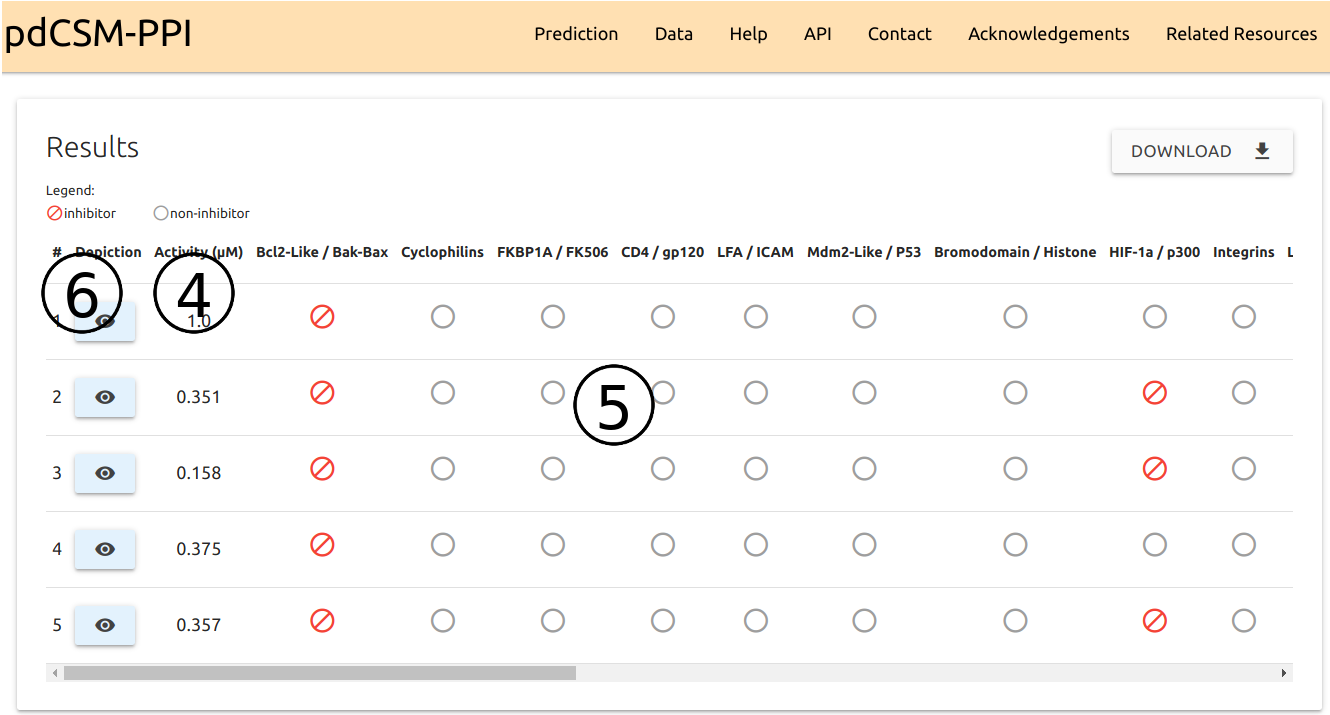
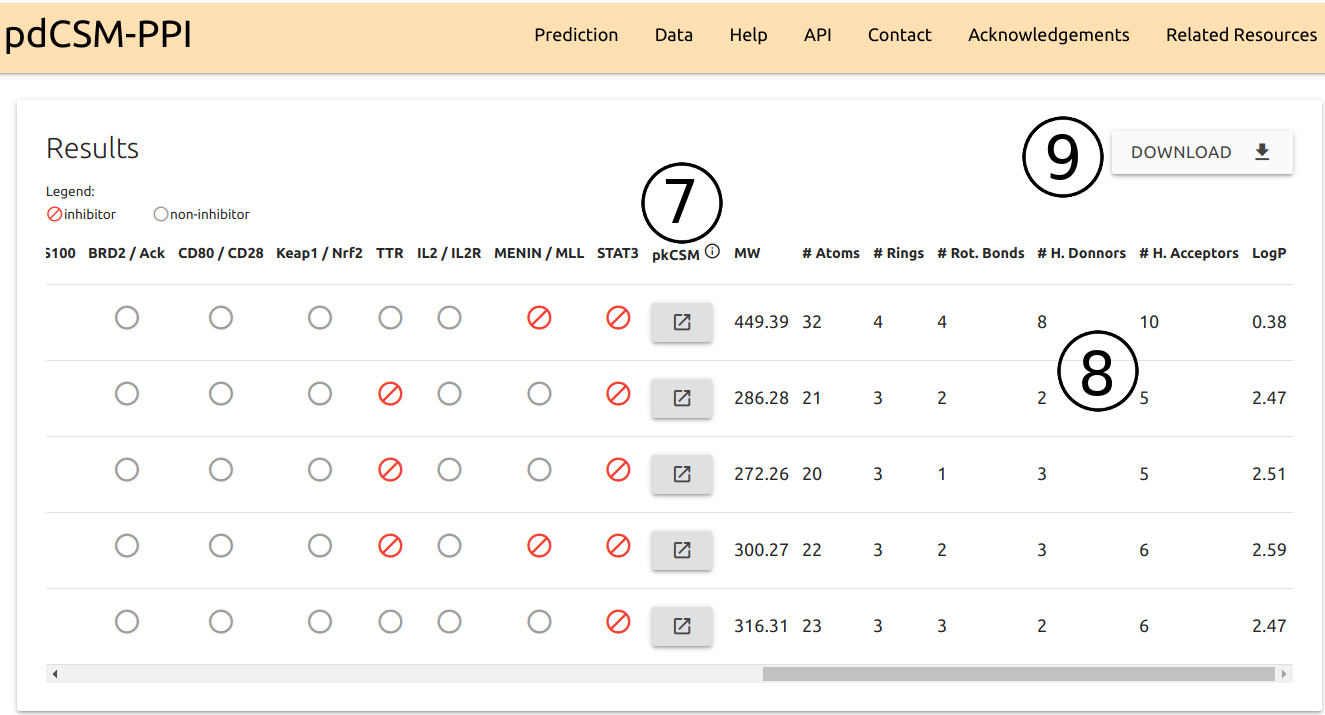
Results

For both input options (single and list), results are summarised as a downloadable table with the following information:
- PPI inhibitory potency predictions (IC50), in µM, for each compound provided (4), as well as class-specific prediction outcomes for 21 different PPI targets (5).
- Small-molecule depiction, generated via SmilesDrawer, can be viewed by clicking the buttons available in (6).
- Buttons for calculating additional pharmacokinetic properties via pkCSM are also available (7).
- General physicochemical properties, including Molecular weight (MW), number of hydrogen donors, acceptors and rings, and LogP (8).
- The entire table can be downloaded as a comma-separated values file (CSV) using the button right above the table (9).

Contact us
In case you experience any troube using pdCSM-PPI or if you have any suggestions or comments, please do not hesitate in cantacting us either via email or via our Group website.
If your are contacting regarding a job submission, please include details such as input information and the job identifier