Platinum
Protein-ligand affinity change upon mutation database

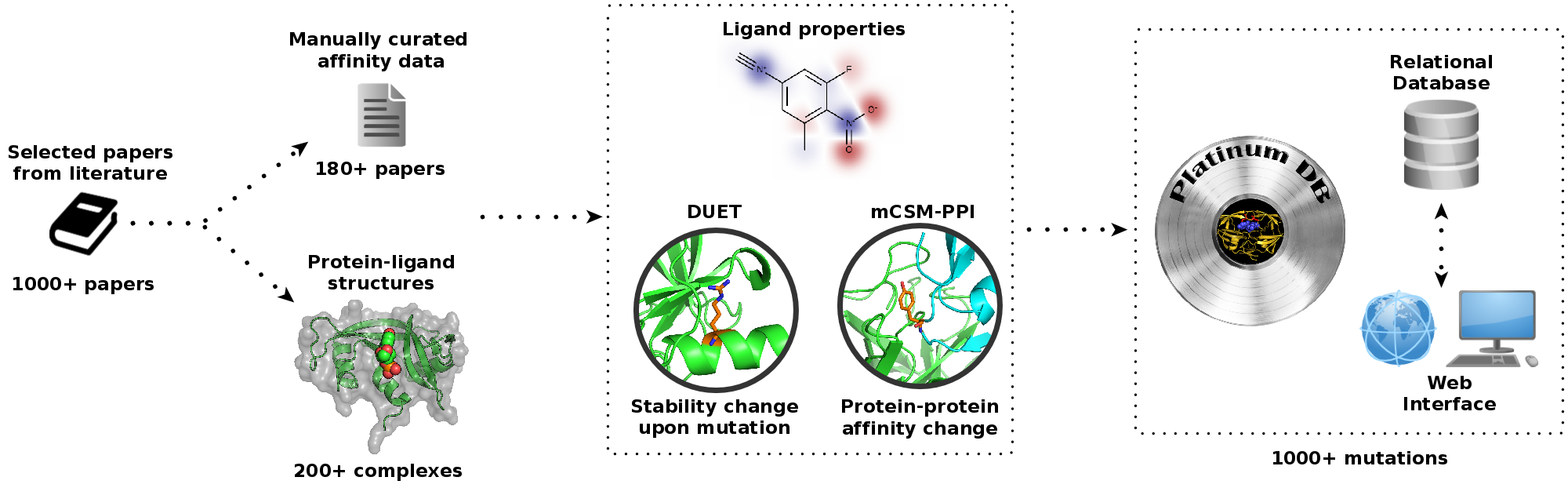
Platinum: a structural database of experimentally measured effects of mutations on protein-ligand complexes
Douglas E. V. Pires, Tom L. Blundell, David B. AscherNucleic Acids Research (Database Issue), v. 43 (D1), p. D387-D391, 2015.


High throughput sequencing initiatives are generating extensive data on non-synonymous single
nucleotide polymorphisms (nsSNPs) in human and other genomes. The strong selective pressure imposed by
small molecule drugs on many quickly evolving systems, including viruses, bacteria and human cancer, can
cause the rapid development of resistance to these therapies.
In order to study and understand the impacts of missense mutations on the interaction of ligands with the
proteome, as well as to guide protein engineering, we have developed Platinum. This manually curated,
literature-derived database comprising over 1,000 mutations for the first time associates experimental
information on changes in protein-ligand affinity with the three-dimensional structures of the complex.
To minimise differences arising from experimental techniques and to be able to compare directly binding
affinities, Platinum considers only changes measured by the same group and with the same amino-acid sequence
used for structure determination, providing a direct link between protein structure, how a ligand binds and
how mutations alter the affinity of the ligand for the protein.
We believe that Platinum will be an invaluable resource for understanding the effects of mutations that give
rise to drug resistance, a major problem emerging in pandemics such those caused by the influenza virus, in
infectious diseases such tuberculosis, in cancer and in many other life threatening illnesses.